8K8X
 
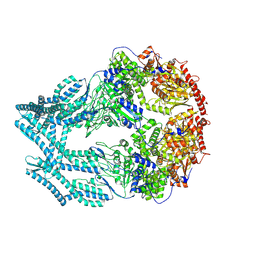 | | CryoEM of LonC open pentamer, apo state | | Descriptor: | endopeptidase La | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
8K94
 
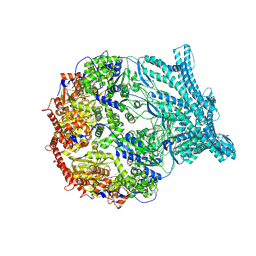 | | CryoEM structure of LonC protease S582A open pentamer with lysozyme | | Descriptor: | Endopeptidase La, Monothiophosphate | | Authors: | Li, M, Hsieh, K, Liu, H, Zhang, S, Gao, Y, Gong, Q, Zhang, K, Chang, C, Li, S. | | Deposit date: | 2023-07-31 | | Release date: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Bifurcated assembly pathway and dual function of a Lon-like protease revealed by cryo-EM Analysis
Fundam Res, 2024
|
|
6A50
 
 | | structure of benzoylformate decarboxylases in complex with cofactor TPP | | Descriptor: | MAGNESIUM ION, THIAMINE DIPHOSPHATE, benzoylformate decarboxylases | | Authors: | Guo, Y, Wang, S, Nie, Y, Li, S. | | Deposit date: | 2018-06-21 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Synthetic Pathway for Acetyl-Coenzyme A Biosynthesis
Nat Commun, 2019
|
|
8IV5
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 1C4 (local refinement) | | Descriptor: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV8
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 3E2 and 1C4 (local refinement) | | Descriptor: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IVA
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs XMA01 and 3E2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV4
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 3E2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
4BLG
 
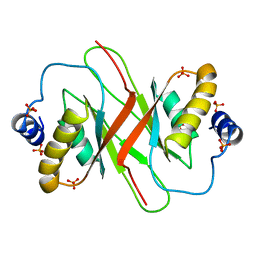 | | Crystal structure of MHV-68 Latency-associated nuclear antigen (LANA) C-terminal DNA binding domain | | Descriptor: | LATENCY-ASSOCIATED NUCLEAR ANTIGEN, PHOSPHATE ION | | Authors: | Correia, B, Cerqueira, S.A, Beauchemin, C, Pires De Miranda, M, Li, S, Ponnusamy, R, Rodrigues, L, Schneider, T.R, Carrondo, M.A, Kaye, K.M, Simas, J.P, McVey, C.E. | | Deposit date: | 2013-05-02 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Gamma-2 Herpesvirus Lana DNA Binding Domain Identifies Charged Surface Residues which Impact Viral Latency
Plos Pathog., 9, 2013
|
|
3QE7
 
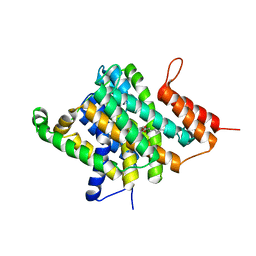 | | Crystal Structure of Uracil Transporter--UraA | | Descriptor: | URACIL, Uracil permease, nonyl beta-D-glucopyranoside | | Authors: | Lu, F.R, Li, S, Yan, N. | | Deposit date: | 2011-01-20 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.781 Å) | | Cite: | Structure and mechanism of the uracil transporter UraA
Nature, 472, 2011
|
|
6V5C
 
 | | Human Drosha and DGCR8 in complex with Primary MicroRNA (MP/RNA complex) - partially docked state | | Descriptor: | Microprocessor complex subunit DGCR8, Pri-miR-16-2 (66-MER), Ribonuclease 3 | | Authors: | Partin, A, Zhang, K, Jeong, B, Herrell, E, Li, S, Chiu, W, Nam, Y. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA.
Mol.Cell, 78, 2020
|
|
2AWT
 
 | | Solution Structure of Human Small Ubiquitin-Like Modifier Protein Isoform 2 (SUMO-2) | | Descriptor: | Small ubiquitin-related modifier 2 | | Authors: | Chang, C.K, Wang, Y.H, Chung, T.L, Chang, C.F, Li, S.S.L, Huang, T.H. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Human Small Ubiquitin-Like Modifier Protein Isoform 2 (SUMO-2)
To be Published
|
|
6V5B
 
 | | Human Drosha and DGCR8 in complex with Primary MicroRNA (MP/RNA complex) - Active state | | Descriptor: | CALCIUM ION, Microprocessor complex subunit DGCR8, Pri-miR-16-2 (78-MER), ... | | Authors: | Partin, A, Zhang, K, Jeong, B, Herrell, E, Li, S, Chiu, W, Nam, Y. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA.
Mol.Cell, 78, 2020
|
|
1YOP
 
 | | The solution structure of Kti11p | | Descriptor: | Kti11p, ZINC ION | | Authors: | Sun, J, Zhang, J, Wu, F, Xu, C, Li, S, Zhao, W, Wu, Z, Wu, J, Zhou, C.-Z, Shi, Y. | | Deposit date: | 2005-01-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Kti11p from Saccharomyces cerevisiae reveals a novel zinc-binding module.
Biochemistry, 44, 2005
|
|
6O3T
 
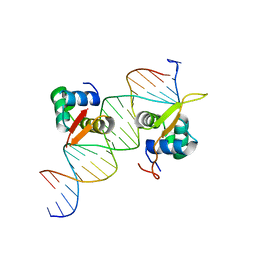 | | Structural basis of FOXC2 and DNA interactions | | Descriptor: | DNA (5'-D(*AP*AP*AP*TP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*GP*CP*CP*CP*G)-3'), DNA (5'-D(*TP*TP*CP*GP*GP*GP*CP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*AP*T)-3'), Forkhead box protein C2 | | Authors: | Nam, H.-J, Li, S. | | Deposit date: | 2019-02-27 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Crystal Structure of FOXC2 in Complex with DNA Target.
Acs Omega, 4, 2019
|
|
6WLT
 
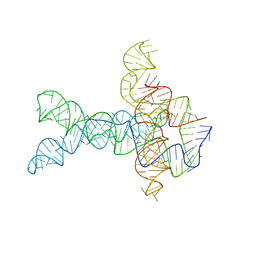 | | Apo V. cholerae glycine riboswitch models, 4.8 Angstrom resolution | | Descriptor: | RNA (231-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLJ
 
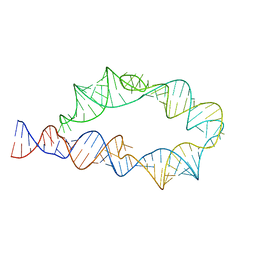 | | ATP-TTR-3 with AMP models, 9.6 Angstrom resolution | | Descriptor: | RNA (130-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLL
 
 | | Apo F. nucleatum glycine riboswitch models, 10.0 Angstrom resolution | | Descriptor: | RNA (171-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLQ
 
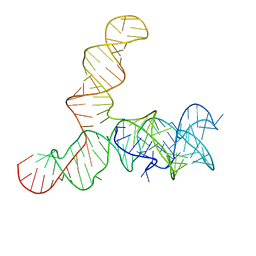 | | Apo SAM-IV riboswitch models, 4.7 Angstrom resolution | | Descriptor: | RNA (119-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLO
 
 | | hc16 ligase models, 11.0 Angstrom resolution | | Descriptor: | RNA (338-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLN
 
 | | hc16 ligase product models, 10.0 Angstrom resolution | | Descriptor: | RNA (349-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLM
 
 | | F. nucleatum glycine riboswitch with glycine models, 7.4 Angstrom resolution | | Descriptor: | RNA (171-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLR
 
 | | SAM-IV riboswitch with SAM models, 4.8 Angstrom resolution | | Descriptor: | RNA (119-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6UES
 
 | | Apo SAM-IV Riboswitch | | Descriptor: | RNA (119-MER) | | Authors: | Zhang, K, Li, S, Kappel, K, Pintilie, G, Su, Z, Mou, T, Schmid, M, Das, R, Chiu, W. | | Deposit date: | 2019-09-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 angstrom resolution.
Nat Commun, 10, 2019
|
|
6UET
 
 | | SAM-bound SAM-IV riboswitch | | Descriptor: | RNA (119-MER), S-ADENOSYLMETHIONINE | | Authors: | Zhang, K, Li, S, Kappel, K, Pintilie, G, Su, Z, Mou, T, Schmid, M, Das, R, Chiu, W. | | Deposit date: | 2019-09-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 angstrom resolution.
Nat Commun, 10, 2019
|
|
6WLS
 
 | | Tetrahymena ribozyme models, 6.8 Angstrom resolution | | Descriptor: | RNA (388-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
