7YCH
 
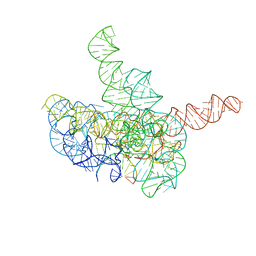 | | Cryo-EM structure of Tetrahymena ribozyme conformation 3 undergoing the first-step self-splicing | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA (393-MER), ... | | Authors: | Zhang, X, Li, S, Pintilie, G, Palo, M.Z, Zhang, K. | | Deposit date: | 2022-07-01 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Snapshots of the first-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8UYM
 
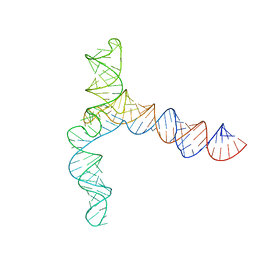 | | MERS 5' proximal stem-loop 5, conformation 3 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYK
 
 | | MERS 5' proximal stem-loop 5, conformation 1 | | Descriptor: | MERS 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYE
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 1 | | Descriptor: | BtCoV-HKU5 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYG
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 2 | | Descriptor: | RNA (135-MER) | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYP
 
 | | SARS-CoV-1 5' proximal stem-loop 5 | | Descriptor: | SARS-CoV-1 5' proximal stem-loop 5 | | Authors: | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | Deposit date: | 2023-11-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2Y08
 
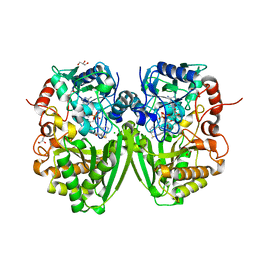 | | Structure of the substrate-free FAD-dependent tirandamycin oxidase TamL | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2010-11-30 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
2Y3R
 
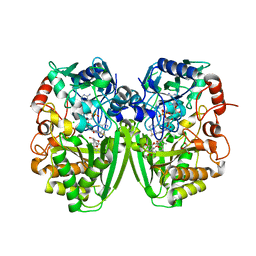 | | Structure of the tirandamycin-bound FAD-dependent tirandamycin oxidase TamL in P21 space group | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2010-12-22 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
7KIP
 
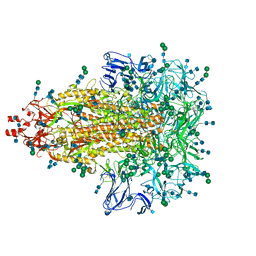 | | A 3.4 Angstrom cryo-EM structure of the human coronavirus spike trimer computationally derived from vitrified NL63 virus particles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhang, K, Li, S, Pintilie, G, Chmielewski, D, Schmid, M, Simmons, G, Jin, J, Chiu, W. | | Deposit date: | 2020-10-24 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | A 3.4- angstrom cryo-EM structure of the human coronavirus spike trimer computationally derived from vitrified NL63 virus particles.
Biorxiv, 2020
|
|
3D23
 
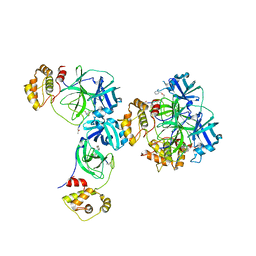 | | Main protease of HCoV-HKU1 | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Zhao, Q, Chen, C, Li, S, Zou, Y. | | Deposit date: | 2008-05-07 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1.
J.Virol., 82, 2008
|
|
3DOF
 
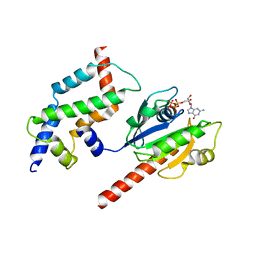 | | Complex of ARL2 and BART, Crystal Form 2 | | Descriptor: | ADP-ribosylation factor-like protein 2, ADP-ribosylation factor-like protein 2-binding protein, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, T, Li, S, Ding, J. | | Deposit date: | 2008-07-04 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the ARL2-GTP-BART complex reveals a novel recognition and binding mode of small GTPase with effector
Structure, 17, 2009
|
|
4YTK
 
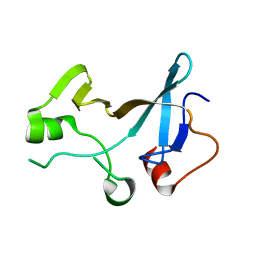 | | Structure of the KOW1-Linker1 domain of Transcription Elongation Factor Spt5 | | Descriptor: | Transcription elongation factor SPT5 | | Authors: | Meyer, P.A, Li, S, Zhang, M, Yamada, K, Takagi, Y, Hartzog, G.A, Fu, J. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.0904 Å) | | Cite: | Structures and Functions of the Multiple KOW Domains of Transcription Elongation Factor Spt5.
Mol.Cell.Biol., 35, 2015
|
|
7L5X
 
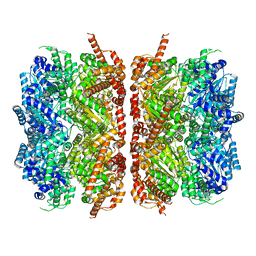 | | p97-R155H mutant dodecamer II | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2020-12-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
7L5W
 
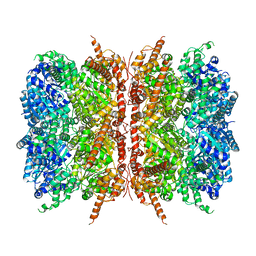 | | p97-R155H mutant dodecamer I | | Descriptor: | Transitional endoplasmic reticulum ATPase | | Authors: | Nandi, P, Li, S, Coulmbres, R.C.A, Wang, F, Williams, D.R, Malyutin, A.G, Poh, Y.-P, Chou, T.-F, Chiu, P.-L. | | Deposit date: | 2020-12-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural and Functional Analysis of Disease-Linked p97 ATPase Mutant Complexes.
Int J Mol Sci, 22, 2021
|
|
4YTL
 
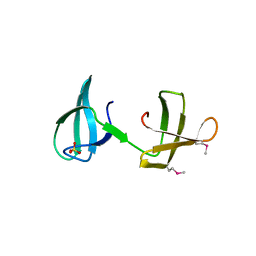 | | Structure of the KOW2-KOW3 Domain of Transcription Elongation Factor Spt5. | | Descriptor: | GLYCEROL, SULFATE ION, Transcription elongation factor SPT5 | | Authors: | Meyer, P.A, Li, S, ZHang, M, Yamada, K, Takagi, Y, Hartzog, G.A, Fu, J. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structures and Functions of the Multiple KOW Domains of Transcription Elongation Factor Spt5.
Mol.Cell.Biol., 35, 2015
|
|
4PED
 
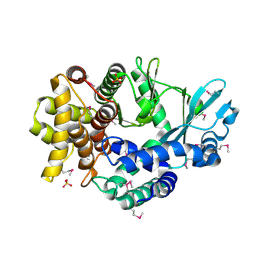 | | Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthes | | Descriptor: | Chaperone activity of bc1 complex-like, mitochondrial, SULFATE ION | | Authors: | Bingman, C.A, Smith, R, Joshi, S, Stefely, J.A, Reidenbach, A.G, Ulbrich, A, Oruganty, O, Floyd, B.J, Jochem, A, Saunders, J.M, Johnson, I.E, Wrobel, R.L, Barber, G.E, Lee, D, Li, S, Kannan, N, Coon, J.J, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mitochondrial ADCK3 Employs an Atypical Protein Kinase-like Fold to Enable Coenzyme Q Biosynthesis.
Mol.Cell, 57, 2015
|
|
2Y3S
 
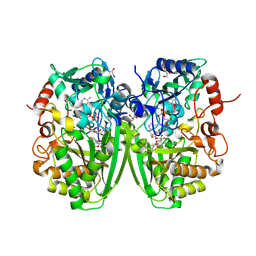 | | Structure of the tirandamycine-bound FAD-dependent tirandamycin oxidase TamL in C2 space group | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2010-12-23 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
2Y4G
 
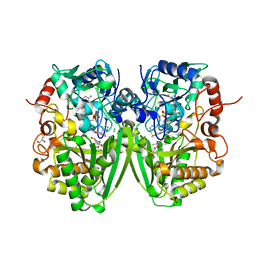 | | Structure of the Tirandamycin-bound FAD-dependent tirandamycin oxidase TamL in P212121 space group | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2011-01-05 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
3DOE
 
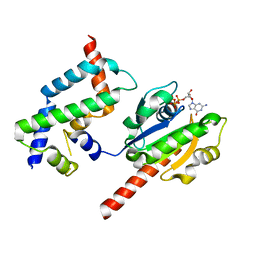 | | Complex of ARL2 and BART, Crystal Form 1 | | Descriptor: | ADP-ribosylation factor-like protein 2, ADP-ribosylation factor-like protein 2-binding protein, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, T, Li, S, Ding, J. | | Deposit date: | 2008-07-04 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the ARL2-GTP-BART complex reveals a novel recognition and binding mode of small GTPase with effector
Structure, 17, 2009
|
|
7YTY
 
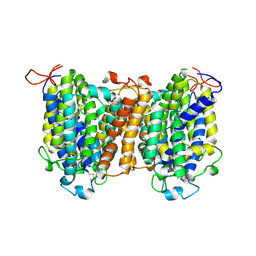 | | Mouse SVCT1 in an apo state | | Descriptor: | Solute carrier family 23 member 1 | | Authors: | She, J, Wang, M, He, J, Zhang, K, Li, S. | | Deposit date: | 2022-08-16 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of vitamin C recognition and transport by mammalian SVCT1 transporter.
Nat Commun, 14, 2023
|
|
2ZIH
 
 | |
7YTW
 
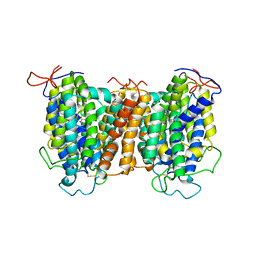 | | Structural basis of vitamin C recognition and transport by mammalian SVCT1 transporter | | Descriptor: | ASCORBIC ACID, SODIUM ION, Solute carrier family 23 member 1 | | Authors: | She, J, Wang, M, He, J, Zhang, K, Li, S. | | Deposit date: | 2022-08-16 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of vitamin C recognition and transport by mammalian SVCT1 transporter.
Nat Commun, 14, 2023
|
|
7WLT
 
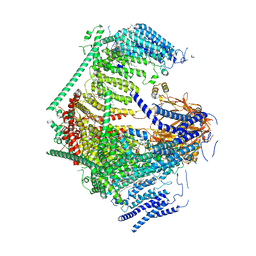 | | the Curved Structure of mPIEZO1 in Lipid Bilayer | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, ... | | Authors: | Yang, X, Lin, C, Chen, X, Li, S, Li, X, Xiao, B. | | Deposit date: | 2022-01-13 | | Release date: | 2022-04-13 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure deformation and curvature sensing of PIEZO1 in lipid membranes.
Nature, 604, 2022
|
|
7WLU
 
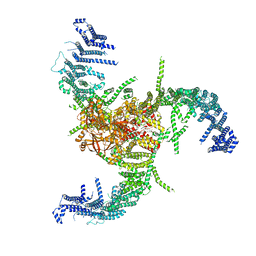 | | The Flattened Structure of mPIEZO1 in Lipid Bilayer | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, Piezo-type mechanosensitive ion channel component 1 | | Authors: | Yang, X, Lin, C, Chen, X, Li, S, Li, X, Xiao, B. | | Deposit date: | 2022-01-13 | | Release date: | 2022-04-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (6.81 Å) | | Cite: | Structure deformation and curvature sensing of PIEZO1 in lipid membranes.
Nature, 604, 2022
|
|
2ZII
 
 | |
