1NB8
 
 | | Structure of the catalytic domain of USP7 (HAUSP) | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Hu, M, Li, P, Li, M, Li, W, Yao, T, Wu, J.-W, Gu, W, Cohen, R.E, Shi, Y. | | Deposit date: | 2002-12-02 | | Release date: | 2003-01-07 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde
Cell(Cambridge,Mass.), 111, 2002
|
|
1NBF
 
 | | Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde | | Descriptor: | Ubiquitin aldehyde, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Hu, M, Li, P, Li, M, Li, W, Yao, T, Wu, J.-W, Gu, W, Cohen, R.E, Shi, Y. | | Deposit date: | 2002-12-02 | | Release date: | 2003-01-07 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde
Cell(Cambridge,Mass.), 111, 2002
|
|
1Z28
 
 | | Crystal Structures of SULT1A2 and SULT1A1*3: Implications in the bioactivation of N-hydroxy-2-acetylamino fluorine (OH-AAF) | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Phenol-sulfating phenol sulfotransferase 1 | | Authors: | Lu, J, Li, H, Liu, M.C, Zhang, J, Li, M, An, X, Chang, W. | | Deposit date: | 2005-03-07 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of SULT1A2 and SULT1A1 *3: insights into the substrate inhibition and the role of Tyr149 in SULT1A2.
Biochem.Biophys.Res.Commun., 396, 2010
|
|
1Z29
 
 | | Crystal Structures of SULT1A2 and SULT1A1*3: Implications in the bioactivation of N-hydroxy-2-acetylamino fluorine (OH-AAF) | | Descriptor: | ACETIC ACID, ADENOSINE-3'-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Lu, J, Li, H, Liu, M.C, Zhang, J, Li, M, An, X, Chang, W. | | Deposit date: | 2005-03-07 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of SULT1A2 and SULT1A1 *3: insights into the substrate inhibition and the role of Tyr149 in SULT1A2.
Biochem.Biophys.Res.Commun., 396, 2010
|
|
2A3R
 
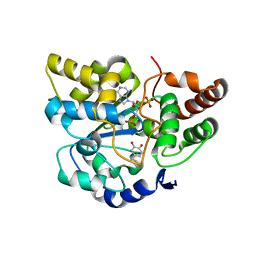 | | Crystal Structure of Human Sulfotransferase SULT1A3 in Complex with Dopamine and 3-Phosphoadenosine 5-Phosphate | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, L-DOPAMINE, Monoamine-sulfating phenol sulfotransferase | | Authors: | Lu, J.H, Li, H.T, Liu, M.C, Zhang, J.P, Li, M, An, X.M, Chang, W.R. | | Deposit date: | 2005-06-26 | | Release date: | 2005-08-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human sulfotransferase SULT1A3 in complex with dopamine and 3'-phosphoadenosine 5'-phosphate
Biochem.Biophys.Res.Commun., 335, 2005
|
|
8R2C
 
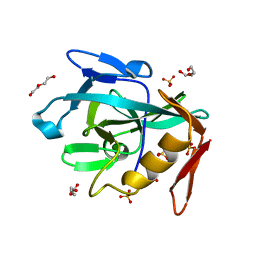 | | Crystal structure of the Vint domain from Tetrahymena thermophila | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, von willebrand factor type A (VWA) domain was originally protein | | Authors: | Iwai, H, Beyer, H.M, Johannson, J.E, Li, M, Wlodawer, A. | | Deposit date: | 2023-11-03 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The three-dimensional structure of the Vint domain from Tetrahymena thermophila suggests a ligand-regulated cleavage mechanism by the HINT fold.
Febs Lett., 598, 2024
|
|
8FNJ
 
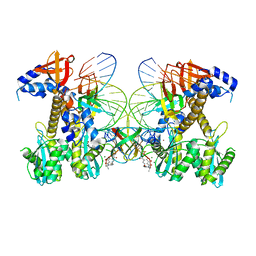 | | Structure of E138K/G140A HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNN
 
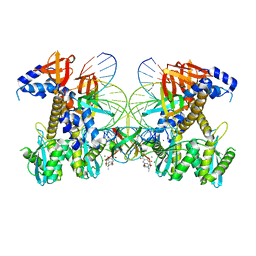 | | Structure of E138K/G140A/Q148K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FN7
 
 | | Structure of WT HIV-1 intasome bound to Dolutegravir | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNL
 
 | | Structure of E138K/Q148K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FND
 
 | | Structure of E138K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNO
 
 | | Structure of E138K/G140A/Q148R HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-28 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNH
 
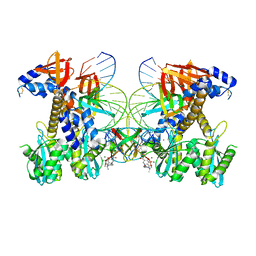 | | Structure of Q148K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNG
 
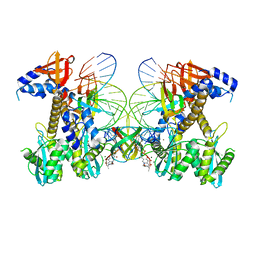 | | Structure of E138K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNP
 
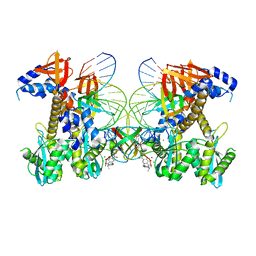 | | Structure of E138K/G140S/Q148H HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNM
 
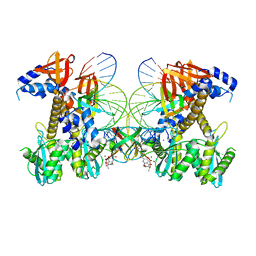 | | Structure of G140A/Q148K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNQ
 
 | | Structure of E138K/G140A/Q148K HIV-1 intasome with 4d bound | | Descriptor: | 4-amino-N-[(2,4-difluorophenyl)methyl]-1-hydroxy-6-(6-hydroxyhexyl)-2-oxo-1,2-dihydro-1,8-naphthyridine-3-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
1B11
 
 | | STRUCTURE OF FELINE IMMUNODEFICIENCY VIRUS PROTEASE COMPLEXED WITH TL-3-093 | | Descriptor: | PROTEIN (Feline Immunodeficiency Virus PROTEASE), SULFATE ION, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Gustchina, A, Li, M, Wlodawer, A. | | Deposit date: | 1998-11-25 | | Release date: | 1998-12-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural studies of FIV and HIV-1 proteases complexed with an efficient inhibitor of FIV protease
Proteins, 38, 2000
|
|
8W4Q
 
 | | Crystal structure of PDE4D complexed with CX-4945 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8W4R
 
 | | Crystal structure of PDE4D complexed with CVT-313 | | Descriptor: | 1,2-ETHANEDIOL, 2,2'-{[6-{[(4-methoxyphenyl)methyl]amino}-9-(propan-2-yl)-9H-purin-2-yl]azanediyl}di(ethan-1-ol), MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8Z9H
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription elongation-reception conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
1AJ4
 
 | | STRUCTURE OF CALCIUM-SATURATED CARDIAC TROPONIN C, NMR, 1 STRUCTURE | | Descriptor: | CALCIUM ION, TROPONIN C | | Authors: | Sia, S.K, Li, M.X, Spyracopoulos, L, Gagne, S.M, Liu, W, Putkey, J.A, Sykes, B.D. | | Deposit date: | 1997-05-14 | | Release date: | 1998-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of cardiac muscle troponin C unexpectedly reveals a closed regulatory domain.
J.Biol.Chem., 272, 1997
|
|
1B8Q
 
 | | SOLUTION STRUCTURE OF THE EXTENDED NEURONAL NITRIC OXIDE SYNTHASE PDZ DOMAIN COMPLEXED WITH AN ASSOCIATED PEPTIDE | | Descriptor: | PROTEIN (HEPTAPEPTIDE), PROTEIN (NEURONAL NITRIC OXIDE SYNTHASE) | | Authors: | Tochio, H, Zhang, Q, Mandal, P, Li, M, Zhang, M. | | Deposit date: | 1999-02-01 | | Release date: | 1999-04-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the extended neuronal nitric oxide synthase PDZ domain complexed with an associated peptide.
Nat.Struct.Biol., 6, 1999
|
|
8W4S
 
 | | Crystal structure of PDE5A in complex with CVT-313 | | Descriptor: | 2,2'-{[6-{[(4-methoxyphenyl)methyl]amino}-9-(propan-2-yl)-9H-purin-2-yl]azanediyl}di(ethan-1-ol), MAGNESIUM ION, ZINC ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8W4T
 
 | | Crystal structure of PDE5A in complex with a novel inhibitor | | Descriptor: | 2-[bis(2-hydroxyethyl)amino]-6-[(4-methoxyphenyl)methylamino]-9-propan-2-yl-7~{H}-purin-8-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
