8IG7
 
 | |
8IG9
 
 | |
8IGA
 
 | |
8IM6
 
 | | Crystal structure of HCoV 229E main protease in complex with PF07304814 | | Descriptor: | 3C-like proteinase, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhou, Y.R, Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2023-03-06 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis of main proteases of HCoV-229E bound to inhibitor PF-07304814 and PF-07321332.
Biochem.Biophys.Res.Commun., 657, 2023
|
|
8IPT
 
 | |
8IPR
 
 | |
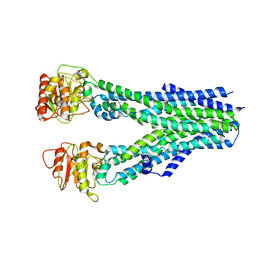
8IPS
 
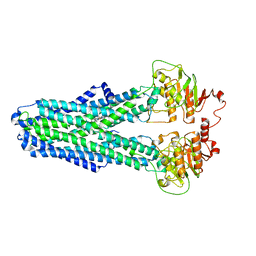 | |
8IPQ
 
 | |
1BWP
 
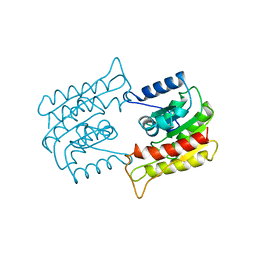 | | PROBING THE SUBSTRATE SPECIFICITY OF THE INTRACELLULAR BRAIN PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Descriptor: | PLATELET-ACTIVATING FACTOR ACETYLHYDROLASE | | Authors: | Ho, Y.S, Sheffield, P.J, Masuyama, J, Arai, H, Li, J, Aoki, J, Inoue, K, Derewenda, U, Derewenda, Z. | | Deposit date: | 1998-09-27 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the Substrate Specificity of the Intracellular Brain Platelet-Activating Factor Acetylhydrolase
Protein Eng., 12, 1999
|
|
6AEI
 
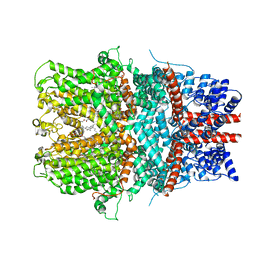 | | Cryo-EM structure of the receptor-activated TRPC5 ion channel | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Duan, J, Li, Z, Li, J, Zhang, J. | | Deposit date: | 2018-08-05 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structure of TRPC5 at 2.8- angstrom resolution reveals unique and conserved structural elements essential for channel function.
Sci Adv, 5, 2019
|
|
4I42
 
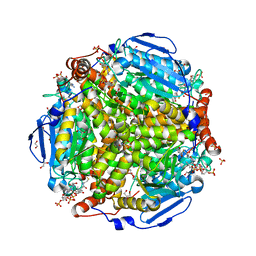 | | E.coli. 1,4-dihydroxy-2-naphthoyl coenzyme A synthase (ecMenB) in complex with 1-hydroxy-2-naphthoyl-CoA | | Descriptor: | 1,2-ETHANEDIOL, 1,4-Dihydroxy-2-naphthoyl-CoA synthase, 1-hydroxy-2-naphthoyl-CoA, ... | | Authors: | Sun, Y, Song, H, Li, J, Li, Y, Jiang, M, Zhou, J, Guo, Z. | | Deposit date: | 2012-11-27 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme A synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
4I52
 
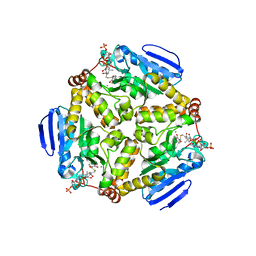 | | scMenB im complex with 1-hydroxy-2-naphthoyl-CoA | | Descriptor: | 1-hydroxy-2-naphthoyl-CoA, CHLORIDE ION, Naphthoate synthase | | Authors: | Song, H.G, Sun, Y.R, Li, J, Li, Y, Jiang, M, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme a synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
4I4Z
 
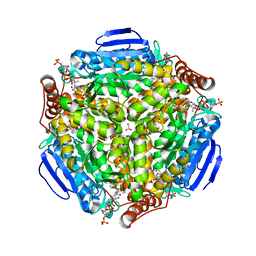 | | Synechocystis sp. PCC 6803 1,4-dihydroxy-2-naphthoyl-coenzyme A synthase (MenB) in complex with salicylyl-CoA | | Descriptor: | BICARBONATE ION, MALONATE ION, Naphthoate synthase, ... | | Authors: | Song, H.G, Sun, Y.R, Li, J, Li, Y, Jiang, M, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme a synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
2KHT
 
 | | NMR Structure of human alpha defensin HNP-1 | | Descriptor: | Neutrophil defensin 1 | | Authors: | Zhang, Y, Li, S, Doherty, T.F, Lubkowski, J, Lu, W, Li, J, Barinka, C, Hong, M. | | Deposit date: | 2009-04-11 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Resonance assignment and three-dimensional structure determination of a human alpha-defensin, HNP-1, by solid-state NMR.
J.Mol.Biol., 397, 2010
|
|
7YFU
 
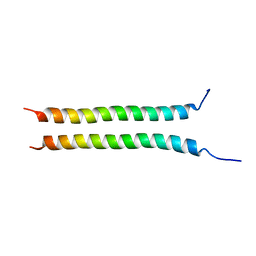 | | Structure of Rpgrip1l CC2 | | Descriptor: | Protein fantom | | Authors: | He, R, Chen, G, Li, Z, Li, J. | | Deposit date: | 2022-07-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the N-terminal coiled-coil domains of the ciliary protein Rpgrip1l.
Iscience, 26, 2023
|
|
6IZQ
 
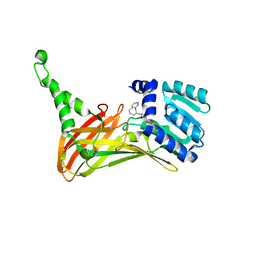 | | PRMT4 bound with a bicyclic compound | | Descriptor: | (2R)-1-(methylamino)-3-(1,3,4,5-tetrahydro-2-benzazepin-2-yl)propan-2-ol, Histone-arginine methyltransferase CARM1 | | Authors: | Xiong, B, Cao, D.Y, Guo, Z.H, Li, Y.L, Li, J, Huang, X, Shen, J.K. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Design and Synthesis of Potent, Selective Inhibitors of Protein Arginine Methyltransferase 4 against Acute Myeloid Leukemia.
J.Med.Chem., 62, 2019
|
|
7YFV
 
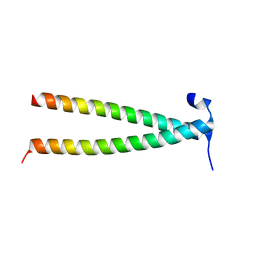 | | Structure of Rpgrip1l CC1 | | Descriptor: | Protein fantom | | Authors: | He, R, Chen, G, Li, Z, Li, J. | | Deposit date: | 2022-07-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the N-terminal coiled-coil domains of the ciliary protein Rpgrip1l.
Iscience, 26, 2023
|
|
8HQI
 
 | |
8W7J
 
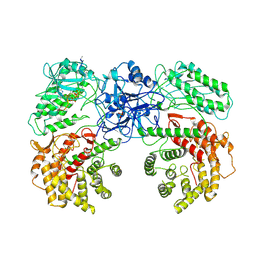 | | Cryo-EM structure of ClassIII Lanthipeptide modification enzyme PneKC with chain A bounded to substrate PneA and GTP. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, PneA LP, ... | | Authors: | Li, Y, Luo, M, Shao, K, Li, J, Li, Z. | | Deposit date: | 2023-08-30 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Mechanistic insights into lanthipeptide modification by a distinct subclass of LanKC enzyme that forms dimers.
Nat Commun, 15, 2024
|
|
8FNE
 
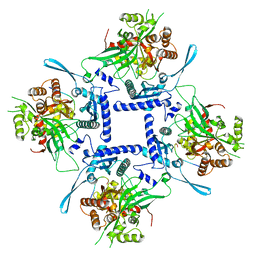 | | phiPA3 PhuN Tetramer, p2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein, PhuN | | Authors: | Nieweglowska, E.S, Brilot, A.F, Mendez-Moran, M, Kokontis, C, Baek, M, Li, J, Cheng, Y, Baker, D, Bondy-Denomy, J, Agard, D.A. | | Deposit date: | 2022-12-27 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The phi PA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice.
Nat Commun, 14, 2023
|
|
8FV5
 
 | | Representation of 16-mer phiPA3 PhuN Lattice, p2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein, phiPA3 PhuN | | Authors: | Nieweglowska, E.S, Brilot, A.F, Mendez-Moran, M, Kokontis, C, Baek, M, Li, J, Cheng, Y, Baker, D, Bondy-Denomy, J, Agard, D.A. | | Deposit date: | 2023-01-18 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.21 Å) | | Cite: | The phi PA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice.
Nat Commun, 14, 2023
|
|
8HVN
 
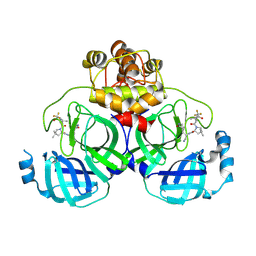 | | Crystal structure of SARS-Cov-2 main protease P132H mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Li, W.W, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
P132H mutant in complex with PF07321332
To Be Published
|
|
8D3C
 
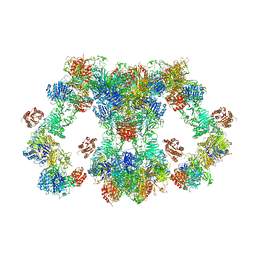 | | VWF tubule derived from monomeric D1-A1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand factor | | Authors: | Anderson, J.R, Li, J, Springer, T.A, Brown, A. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of VWF tubules before and after concatemerization reveal a mechanism of disulfide bond exchange.
Blood, 140, 2022
|
|
8D3D
 
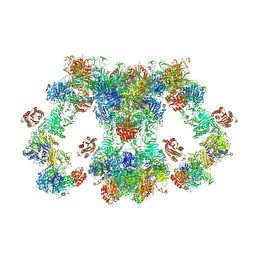 | | VWF tubule derived from dimeric D1-A1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, von Willebrand factor | | Authors: | Anderson, J.R, Li, J, Springer, T.A, Brown, A. | | Deposit date: | 2022-06-01 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of VWF tubules before and after concatemerization reveal a mechanism of disulfide bond exchange.
Blood, 140, 2022
|
|
8C54
 
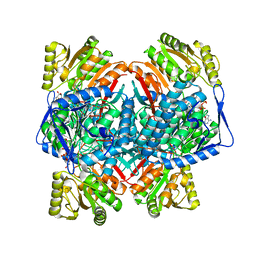 | | Cryo-EM structure of NADH bound SLA dehydrogenase RlGabD from Rhizobium leguminosarum bv. trifolii SRD1565 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Succinate semialdehyde dehydrogenase | | Authors: | Sharma, M, Meek, R.W, Armstrong, Z, Blaza, J.N, Alhifthi, A, Li, J, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2023-01-06 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Molecular basis of sulfolactate synthesis by sulfolactaldehyde dehydrogenase from Rhizobium leguminosarum.
Chem Sci, 14, 2023
|
|
