7C2Y
 
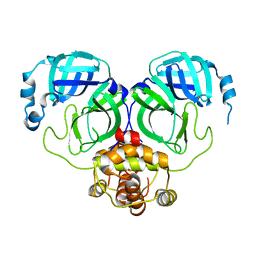 | | The crystal structure of COVID-2019 main protease in the apo state | | Descriptor: | 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zhou, H, Hu, X.H, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-05-10 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | COVID-2019 main protease in the apo state
To Be Published
|
|
2KRG
 
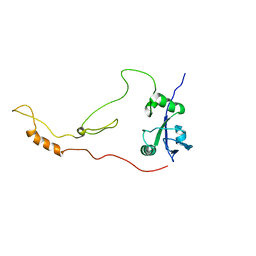 | | Solution Structure of human sodium/ hydrogen exchange regulatory factor 1(150-358) | | Descriptor: | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | | Authors: | Bhattacharya, S, Dai, Z, Li, J, Baxter, S, Callaway, D.J.E, Cowburn, D, Bu, Z. | | Deposit date: | 2009-12-17 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A conformational switch in the scaffolding protein NHERF1 controls autoinhibition and complex formation.
J.Biol.Chem., 285, 2010
|
|
3UOT
 
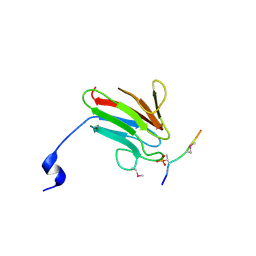 | | Crystal Structure of MDC1 FHA Domain in Complex with a Phosphorylated Peptide from the MDC1 N-terminus | | Descriptor: | Mediator of DNA damage checkpoint protein 1 | | Authors: | Clapperton, J.A, Lloyd, J, Haire, L.F, Li, J, Smerdon, S.J. | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-28 | | Last modified: | 2012-07-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis of ATM-dependent dimerization of the Mdc1 DNA damage checkpoint mediator.
Nucleic Acids Res., 40, 2012
|
|
7CCD
 
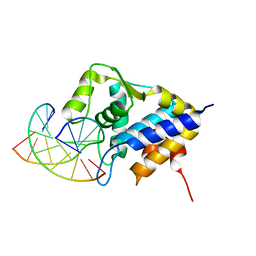 | | Sulfur binding domain of SprMcrA complexed with phosphorothioated DNA | | Descriptor: | DNA (5'-D(*CP*AP*CP*GP*TP*TP*CP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*GP*AS*AP*CP*GP*TP*G)-3'), HNHc domain-containing protein | | Authors: | Yu, H, Li, J, Liu, G, Zhao, G, Wang, Y, Hu, W, Deng, Z, Gan, J, Zhao, Y, He, X. | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | DNA backbone interactions impact the sequence specificity of DNA sulfur-binding domains: revelations from structural analyses.
Nucleic Acids Res., 48, 2020
|
|
7CBY
 
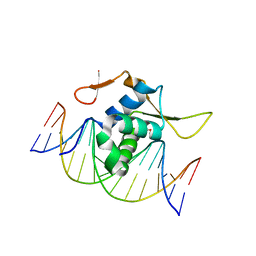 | | Structure of FOXG1 DNA binding domain bound to DBE2 DNA site | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), ... | | Authors: | Dai, S.Y, Li, J, Chen, Y.H. | | Deposit date: | 2020-06-15 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.646 Å) | | Cite: | Structural Basis for DNA Recognition by FOXG1 and the Characterization of Disease-causing FOXG1 Mutations.
J.Mol.Biol., 432, 2020
|
|
3N2Y
 
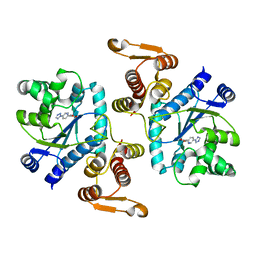 | |
8W7A
 
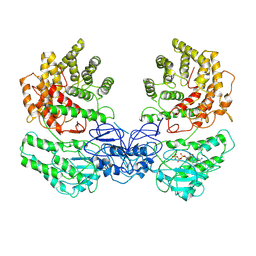 | | Cryo-EM structure of ClassIII Lanthipeptide modification enzyme PneKC in the presence of GTP. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Protein kinase domain-containing protein | | Authors: | Li, Y, Luo, M, Shao, K, Li, J. | | Deposit date: | 2023-08-30 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Mechanistic insights into lanthipeptide modification by a distinct subclass of LanKC enzyme that forms dimers.
Nat Commun, 15, 2024
|
|
8W7J
 
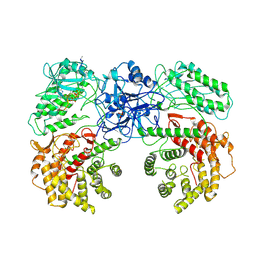 | | Cryo-EM structure of ClassIII Lanthipeptide modification enzyme PneKC with chain A bounded to substrate PneA and GTP. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, PneA LP, ... | | Authors: | Li, Y, Luo, M, Shao, K, Li, J, Li, Z. | | Deposit date: | 2023-08-30 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Mechanistic insights into lanthipeptide modification by a distinct subclass of LanKC enzyme that forms dimers.
Nat Commun, 15, 2024
|
|
8WGO
 
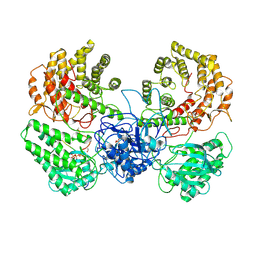 | | Cryo-EM structure of ClassIII Lanthipeptide modification enzyme PneKC in the presence of PneA and GTPrS. | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Li, Y, Luo, M, Shao, K, Li, J. | | Deposit date: | 2023-09-22 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanistic insights into lanthipeptide modification by a distinct subclass of LanKC enzyme that forms dimers.
Nat Commun, 15, 2024
|
|
6KF5
 
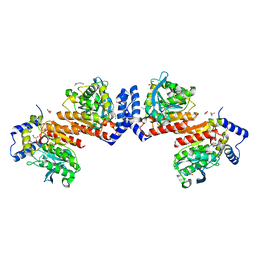 | | Microbial Hormone-sensitive lipase E53 mutant I256L | | Descriptor: | (4-nitrophenyl) hexanoate, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Yang, X, Li, Z.Y, Li, J, Xu, X.W. | | Deposit date: | 2019-07-06 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Microbial Hormone-sensitive lipase E53 mutant I256L
To Be Published
|
|
3RHZ
 
 | |
4QBO
 
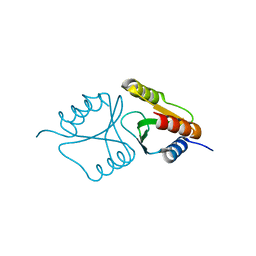 | | VRR_NUC domain | | Descriptor: | MAGNESIUM ION, Nuclease | | Authors: | Smerdon, S.J, Pennell, S, Li, J. | | Deposit date: | 2014-05-08 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | FAN1 activity on asymmetric repair intermediates is mediated by an atypical monomeric virus-type replication-repair nuclease domain.
Cell Rep, 8, 2014
|
|
6KOE
 
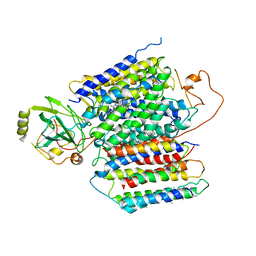 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis | | Descriptor: | 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, ... | | Authors: | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KOC
 
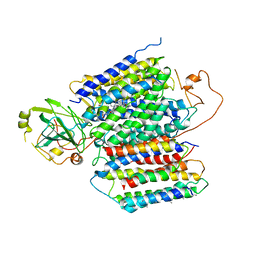 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis complexed with 3-iodo-N-oxo-2-heptyl-4-hydroxyquinoline | | Descriptor: | 2-heptyl-3-iodanyl-1-oxidanyl-quinolin-4-one, AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, ... | | Authors: | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4GHT
 
 | | Crystal structure of EV71 3C proteinase in complex with AG7088 | | Descriptor: | 3C proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4G5S
 
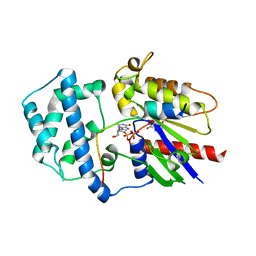 | | Structure of LGN GL3/Galphai3 complex | | Descriptor: | CITRIC ACID, G-protein-signaling modulator 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Jia, M, Li, J, Zhu, J, Wen, W, Zhang, M, Wang, W. | | Deposit date: | 2012-07-18 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Crystal Structures of the scaffolding protein LGN reveal the general mechanism by which GoLoco binding motifs inhibit the release of GDP from Galphai subunits in G-coupled heterotrimeric proteins
To be Published
|
|
2LXH
 
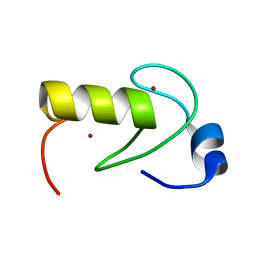 | | NMR structure of the RING domain in ubiquitin ligase gp78 | | Descriptor: | E3 ubiquitin-protein ligase AMFR, ZINC ION | | Authors: | Das, R, Linag, Y, Mariano, J, Li, J, Huang, T, King, A, Weissman, A, Ji, X, Byrd, R. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
6KOB
 
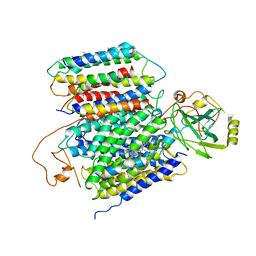 | | X-ray Structure of the proton-pumping cytochrome aa3-600 menaquinol oxidase from Bacillus subtilis | | Descriptor: | AA3-600 quinol oxidase subunit I, AA3-600 quinol oxidase subunit IIII, AA3-600 quinol oxidase subunit IV,Quinol oxidase subunit 4, ... | | Authors: | Xu, J, Ding, Z, Liu, B, Li, J, Gennis, R.B, Zhu, J. | | Deposit date: | 2019-08-09 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the cytochromeaa3-600 heme-copper menaquinol oxidase bound to inhibitor HQNO shows TM0 is part of the quinol binding site.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4N6E
 
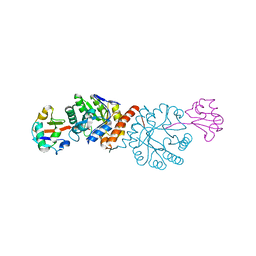 | | Crystal structure of Amycolatopsis orientalis BexX/CysO complex | | Descriptor: | Putative thiosugar synthase, SULFATE ION, ThiS/MoaD family protein | | Authors: | Zhang, X, Zhang, Y, Kinsland, C, Sasaki, E, Sun, H.G, Lu, M.J, Liu, T, Ou, A, Li, J, Chen, Y, Liu, H, Ealick, S.E. | | Deposit date: | 2013-10-11 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Co-opting sulphur-carrier proteins from primary metabolic pathways for 2-thiosugar biosynthesis.
Nature, 509, 2014
|
|
4GHQ
 
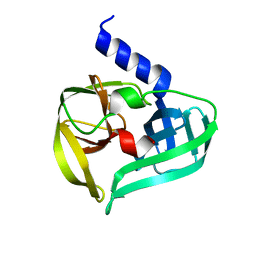 | | Crystal structure of EV71 3C proteinase | | Descriptor: | 3C proteinase | | Authors: | Chen, C, Wu, C, Cai, Q, Li, N, Peng, X, Cai, Y, Yin, K, Chen, X, Wang, X, Zhang, R, Liu, L, Chen, S, Li, J, Lin, T. | | Deposit date: | 2012-08-08 | | Release date: | 2013-06-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Enterovirus 71 3C proteinase (strain E2004104-TW-CDC) and its complex with rupintrivir
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2LXP
 
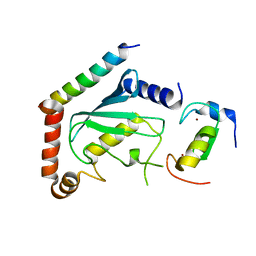 | | NMR structure of two domains in ubiquitin ligase gp78, RING and G2BR, bound to its conjugating enzyme Ube2g | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin-conjugating enzyme E2 G2, ZINC ION | | Authors: | Das, R, Linag, Y, Mariano, J, Li, J, Huang, T, King, A, Weissman, A, Ji, X, Byrd, R. | | Deposit date: | 2012-08-30 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
4NXF
 
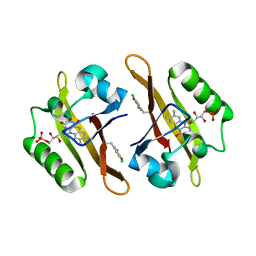 | | Crystal structure of iLOV-I486(2LT) at pH 8.0 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Wang, J, Liu, X, Li, J. | | Deposit date: | 2013-12-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.766 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
8HVU
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
8HVV
 
 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zeng, X.Y, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
8HVX
 
 | | Crystal structure of SARS-Cov-2 main protease Y54C mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
