6WZW
 
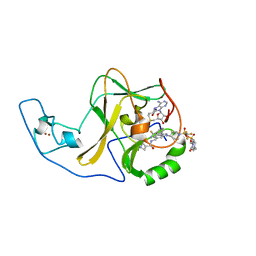 | | Ash1L SET domain in complex with AS-85 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Histone-lysine N-methyltransferase ASH1L, N-{[3-(3-carbamothioylphenyl)-1-{1-[(trifluoromethyl)sulfonyl]piperidin-4-yl}-1H-indol-6-yl]methyl}azetidine-3-carboxamide, ... | | Authors: | Li, H, Deng, J, Cierpicki, T, Grembecka, J. | | Deposit date: | 2020-05-14 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of first-in-class inhibitors of ASH1L histone methyltransferase with anti-leukemic activity.
Nat Commun, 12, 2021
|
|
8F5Z
 
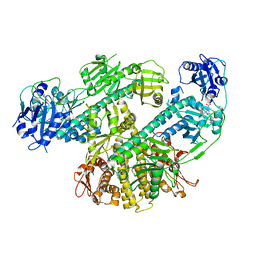 | | Composite map of CryoEM structure of Arabidopsis thaliana phytochrome A | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Li, H, Li, H. | | Deposit date: | 2022-11-15 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The structure of Arabidopsis phytochrome A reveals topological and functional diversification among the plant photoreceptor isoforms.
Nat.Plants, 9, 2023
|
|
7SCJ
 
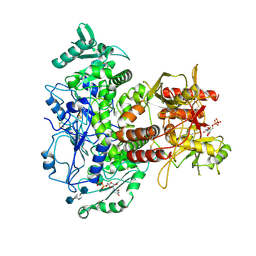 | |
7SCH
 
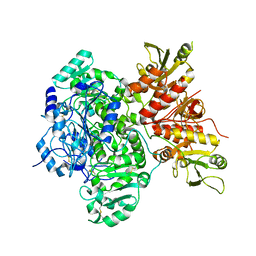 | | Cryo-EM structure of the human Exostosin-1 and Exostosin-2 heterodimer | | Descriptor: | Exostosin-1, Exostosin-2, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, H, Li, H. | | Deposit date: | 2021-09-28 | | Release date: | 2022-09-28 | | Last modified: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for heparan sulfate co-polymerase action by the EXT1-2 complex.
Nat.Chem.Biol., 19, 2023
|
|
2QN5
 
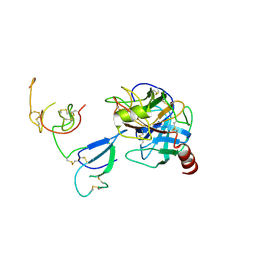 | | Crystal Structure and Functional Study of the Bowman-Birk Inhibitor from Rice Bran in Complex with Bovine Trypsin | | Descriptor: | Bowman-Birk type bran trypsin inhibitor, Cationic trypsin | | Authors: | Li, H.T, Lin, Y.H, Guan, H.H, Hsieh, Y.C, Wang, A.H.J, Chen, C.J. | | Deposit date: | 2007-07-18 | | Release date: | 2008-07-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure and Functional Study of the Bowman-Birk Inhibitor from Rice Bran in Complex with Bovine Trypsin
To be Published
|
|
7RZW
 
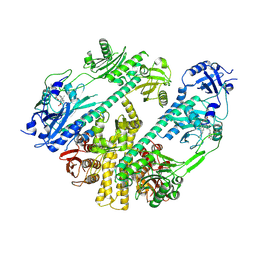 | | CryoEM structure of Arabidopsis thaliana phytochrome B | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome B | | Authors: | Li, H, Burgie, E.S, Vierstra, R.D, Li, H. | | Deposit date: | 2021-08-27 | | Release date: | 2022-04-13 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Plant phytochrome B is an asymmetric dimer with unique signalling potential.
Nature, 604, 2022
|
|
5JLB
 
 | |
6LJG
 
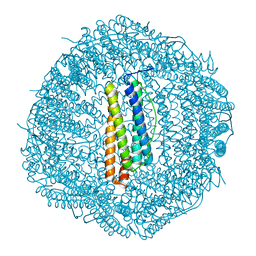 | | Crassostrea gigas ferritin mutant-D119G | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Li, H, Zang, J, Tan, X, Wang, Z, Du, M. | | Deposit date: | 2019-12-15 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crassostrea gigas ferritin mutant-D119G
To Be Published
|
|
6L34
 
 | |
6LIJ
 
 | | Crassostrea gigas ferritin | | Descriptor: | Ferritin, MAGNESIUM ION | | Authors: | Li, H, Zang, J, Tan, X, Wang, Z, Du, M. | | Deposit date: | 2019-12-11 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crassostrea gigas ferritin
To Be Published
|
|
7YI2
 
 | | Cryo-EM structure of Rpd3S in loose-state Rpd3S-NCP complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
8J8I
 
 | |
8J09
 
 | | Crystal structure of protein 3745 | | Descriptor: | Cell division control protein 45, DNA replication regulator SLD3 | | Authors: | Li, H, Yao, M. | | Deposit date: | 2023-04-10 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural and functional insights into a process of complex formation by 3745
To Be Published
|
|
7S05
 
 | | Cryo-EM structure of human GlcNAc-1-phosphotransferase A2B2 subcomplex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, H, Li, H. | | Deposit date: | 2021-08-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the human GlcNAc-1-phosphotransferase alpha beta subunits reveals regulatory mechanism for lysosomal enzyme glycan phosphorylation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S06
 
 | | Cryo-EM structure of human GlcNAc-1-phosphotransferase A2B2 subcomplex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetylglucosamine-1-phosphotransferase subunits alpha/beta | | Authors: | Li, H, Li, H. | | Deposit date: | 2021-08-30 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the human GlcNAc-1-phosphotransferase alpha beta subunits reveals regulatory mechanism for lysosomal enzyme glycan phosphorylation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1FAG
 
 | | STRUCTURE OF CYTOCHROME P450 | | Descriptor: | CYTOCHROME P450 BM-3, PALMITOLEIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, H.Y, Poulos, T.L. | | Deposit date: | 1996-08-01 | | Release date: | 1997-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of the cytochrome p450BM-3 haem domain complexed with the fatty acid substrate, palmitoleic acid.
Nat.Struct.Biol., 4, 1997
|
|
1FV1
 
 | | STRUCTURAL BASIS FOR THE BINDING OF AN IMMUNODOMINANT PEPTIDE FROM MYELIN BASIC PROTEIN IN DIFFERENT REGISTERS BY TWO HLA-DR2 ALLELES | | Descriptor: | GLYCEROL, MAJOR HISTOCOMPATIBILITY COMPLEX ALPHA CHAIN, MAJOR HISTOCOMPATIBILITY COMPLEX BETA CHAIN, ... | | Authors: | Li, H, Mariuzza, A.R, Li, Y, Martin, R. | | Deposit date: | 2000-09-18 | | Release date: | 2000-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the binding of an immunodominant peptide from myelin basic protein in different registers by two HLA-DR2 proteins.
J.Mol.Biol., 304, 2000
|
|
3FP9
 
 | |
2AXH
 
 | | Crystal structures of T cell receptor beta chains related to rheumatoid arthritis | | Descriptor: | T cell receptor beta chain | | Authors: | Li, H, Van Vranken, S, Zhao, Y, Li, Z, Guo, Y, Eisele, L, Li, Y. | | Deposit date: | 2005-09-05 | | Release date: | 2005-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of T cell receptor (beta) chains related to rheumatoid arthritis.
Protein Sci., 14, 2005
|
|
2AXJ
 
 | | Crystal structures of T cell receptor beta chains related to rheumatoid arthritis | | Descriptor: | SF4 T cell receptor beta chain | | Authors: | Li, H, Van Vranken, S, Zhao, Y, Li, Z, Guo, Y, Eisele, L, Li, Y. | | Deposit date: | 2005-09-05 | | Release date: | 2005-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of T cell receptor (beta) chains related to rheumatoid arthritis.
Protein Sci., 14, 2005
|
|
5JLE
 
 | | Crystal structure of SETD2 bound to SAH | | Descriptor: | Histone-lysine N-methyltransferase SETD2, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Li, H, Yang, S, Zheng, X. | | Deposit date: | 2016-04-27 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for oncohistone H3 recognition by SETD2 methyltransferase
Genes Dev., 30, 2016
|
|
7YI3
 
 | | Cryo-EM structure of Rpd3S in close-state Rpd3S-NCP complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI1
 
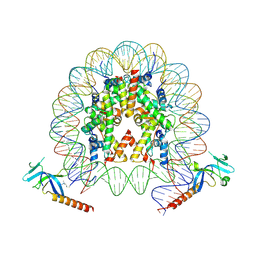 | | Cryo-EM structure of Eaf3 CHD bound to H3K36me3 nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI0
 
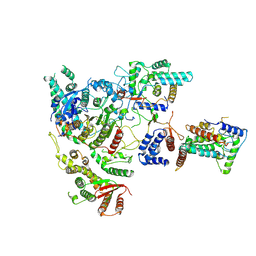 | | Cryo-EM structure of Rpd3S complex | | Descriptor: | Chromatin modification-related protein EAF3, Histone deacetylase RPD3, Transcriptional regulatory protein RCO1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI5
 
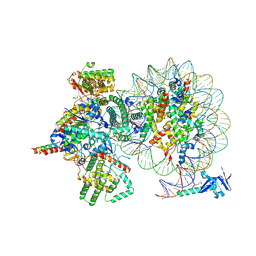 | | Cryo-EM structure of Rpd3S complex bound to H3K36me3 nucleosome in loose state | | Descriptor: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
