6M4S
 
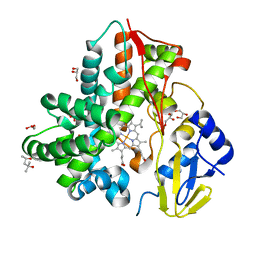 | | Crystal Structure Analysis of the cytochrome P450 CYP-Sb21 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Cytochrome P450 hydroxylase sb21, ... | | Authors: | Li, F.W, Li, S.Y. | | Deposit date: | 2020-03-09 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided manipulation of the regioselectivity of the cyclosporine A hydroxylase CYP-sb21 from Sebekia benihana .
Synth Syst Biotechnol, 5, 2020
|
|
4WHG
 
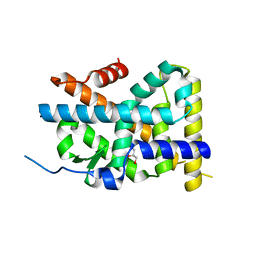 | | Crystal Structure of TR3 LBD in complex with Molecule 3 | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)octan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Wang, Y, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
4WHF
 
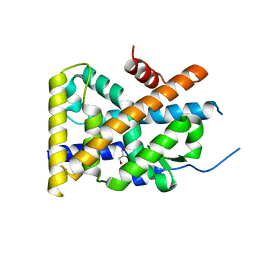 | | Crystal Structure of TR3 LBD in complex with 1-(3,4,5-trihydroxyphenyl)decan-1-one | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)decan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Wang, Y, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
4RE8
 
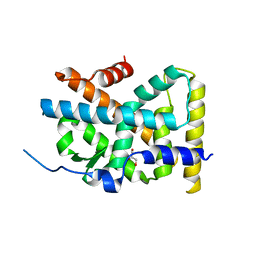 | | Crystal Structure of TR3 LBD in complex with Molecule 5 | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)dodecan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Weijia, W, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
4REE
 
 | | Crystal Structure of TR3 LBD in complex with Molecule 6 | | Descriptor: | 1-(2,3,4-trihydroxyphenyl)nonan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
4REF
 
 | | Crystal Structure of TR3 LBD_L449W in complex with Molecule 2 | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)hexan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
5YSW
 
 | | Crystal Structure Analysis of Rif16 in complex with R-L | | Descriptor: | (2S,12E,14E,16S,17S,18R,19R,20R,21S,22R,23S,24E)-21-(acetyloxy)-5,6,17,19-tetrahydroxy-23-methoxy-2,4,12,16,18,20,22-heptamethyl-1,11-dioxo-1,2-dihydro-2,7-(epoxypentadeca[1,11,13]trienoimino)naphtho[2,1-b]furan-9-yl hydroxyacetate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, F.W, Qi, F.F, Xiao, Y.L, Zhao, G.P, Li, S.Y. | | Deposit date: | 2017-11-15 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Deciphering the late steps of rifamycin biosynthesis.
Nat Commun, 9, 2018
|
|
5YSM
 
 | | Crystal Structure Analysis of Rif16 | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, F.W, Qi, F.F, Xiao, Y.L, Zhao, G.P, Li, S.Y. | | Deposit date: | 2017-11-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Deciphering the late steps of rifamycin biosynthesis.
Nat Commun, 9, 2018
|
|
7Y90
 
 | | Crystal Structure Analysis of cp1 bound BCL2 | | Descriptor: | (2R)-3-[2-(aminomethyl)-3-azanyl-1-[4-[2-(2-chloranylethanoylamino)ethylcarbamoyl]phenyl]prop-1-enyl]sulfanyl-2-(carboxyamino)propanoic acid, Apoptosis regulator Bcl-2, cp1 peptide | | Authors: | Li, F.W. | | Deposit date: | 2022-06-24 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7Y99
 
 | | Crystal Structure Analysis of cp2 bound BCLxl | | Descriptor: | Bcl-2-like protein 1, CP2 peptide, N-(2-acetamidoethyl)-4-(4,5-dihydro-1,3-thiazol-2-yl)benzamide | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-24 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7YAA
 
 | | Crystal structure analysis of cp3 bound BCLxl | | Descriptor: | Bcl-2-like protein 1, GLYCEROL, N-(2-acetamidoethyl)-4-(4-methanoyl-1,3-thiazol-2-yl)benzamide, ... | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7Y8D
 
 | | Crystal structure of cp1 bound BCLxl | | Descriptor: | (2R)-3-[2-(aminomethyl)-3-azanyl-1-[4-[2-(2-chloranylethanoylamino)ethylcarbamoyl]phenyl]prop-1-enyl]sulfanyl-2-(carboxyamino)propanoic acid, Bcl-2-like protein 1, cp1 peptide | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7YA5
 
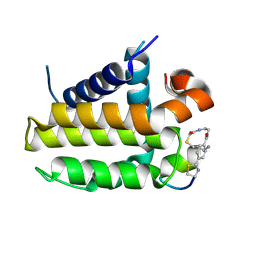 | | Crystal structure analysis of cp1 bound BCL2/G101V | | Descriptor: | (2R)-3-[2-(aminomethyl)-3-azanyl-1-[4-[2-(2-chloranylethanoylamino)ethylcarbamoyl]phenyl]prop-1-enyl]sulfanyl-2-(carboxyamino)propanoic acid, Apoptosis regulator Bcl-2, cp1 peptide | | Authors: | Li, F.W, Liu, C, Wu, C.L, Wu, D.L. | | Deposit date: | 2022-06-27 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7YB7
 
 | | anti-apoptotic protein BCL-2-M12 | | Descriptor: | Apoptosis regulator Bcl-2,Bcl-2-like protein 1, N-(2-acetamidoethyl)-4-(4,5-dihydro-1,3-thiazol-2-yl)benzamide, cp2 peptide | | Authors: | Li, F.W, Liu, C, Wu, D.L. | | Deposit date: | 2022-06-29 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cyclic peptides discriminate BCL-2 and its clinical mutants from BCL-X L by engaging a single-residue discrepancy.
Nat Commun, 15, 2024
|
|
7XI3
 
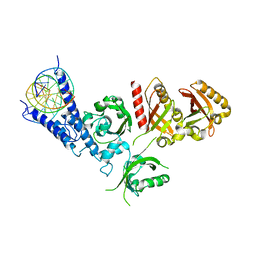 | | Crystal Structure of the NPAS4-ARNT2 heterodimer in complex with DNA | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator 2, DNA (5'-D(P*CP*CP*AP*TP*CP*AP*CP*TP*CP*AP*CP*GP*AP*CP*CP*T)-3'), DNA (5'-D(P*GP*GP*AP*GP*GP*TP*CP*GP*TP*GP*AP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Sun, X.N, Jing, L.Q, Li, F.W, Wu, D.L. | | Deposit date: | 2022-04-11 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.274 Å) | | Cite: | Structures of NPAS4-ARNT and NPAS4-ARNT2 heterodimers reveal new dimerization modalities in the bHLH-PAS transcription factor family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XI4
 
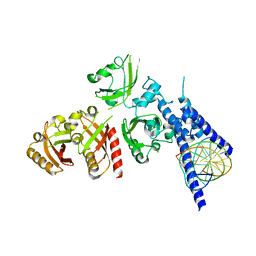 | | Crystal Structure of the NPAS4-ARNT heterodimer in complex with DNA | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, DNA (5'-D(*GP*GP*AP*GP*GP*TP*CP*GP*TP*GP*AP*GP*TP*GP*AP*T)-3'), DNA (5'-D(P*CP*CP*AP*TP*CP*AP*CP*TP*CP*AP*CP*GP*AP*CP*CP*T)-3'), ... | | Authors: | Sun, X.N, Jing, L.Q, Li, F.W, Wu, D.L. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.707 Å) | | Cite: | Structures of NPAS4-ARNT and NPAS4-ARNT2 heterodimers reveal new dimerization modalities in the bHLH-PAS transcription factor family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XHV
 
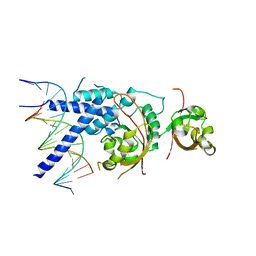 | | Crystal Structure of the NPAS4-ARNT heterodimer in complex with DNA | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, DNA (5'-D(P*CP*CP*AP*TP*CP*AP*CP*TP*CP*AP*CP*GP*AP*CP*CP*T)-3'), DNA (5'-D(P*GP*GP*AP*GP*GP*TP*CP*GP*TP*GP*AP*GP*TP*GP*AP*T)-3'), ... | | Authors: | Sun, X.N, Jing, L.Q, Li, F.W, Wu, D.L. | | Deposit date: | 2022-04-10 | | Release date: | 2022-11-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.996 Å) | | Cite: | Structures of NPAS4-ARNT and NPAS4-ARNT2 heterodimers reveal new dimerization modalities in the bHLH-PAS transcription factor family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7C1L
 
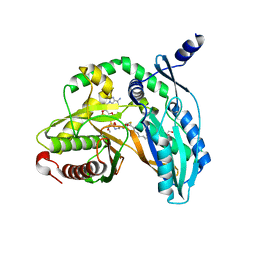 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant R148A in complex with C8-CoA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Non-ribosomal peptide synthetase modules, OCTANOYL-COENZYME A | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1U
 
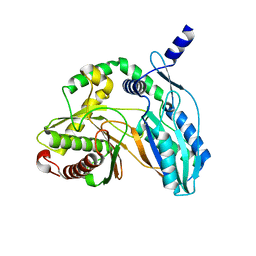 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant H140V/R148A in a "product-released" conformation | | Descriptor: | Non-ribosomal peptide synthetase modules | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1R
 
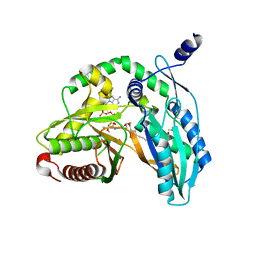 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant H140A/R148A in complex with C8-CoA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Non-ribosomal peptide synthetase modules, OCTANOYL-COENZYME A | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1H
 
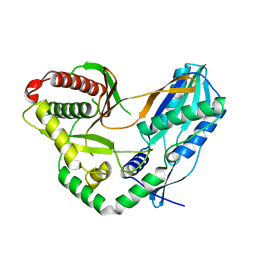 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA | | Descriptor: | Non-ribosomal peptide synthetase modules | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-04 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1P
 
 | | Crystal structure of the starter condensation domain of the rhizomide synthetase RzmA mutant H140V, R148A | | Descriptor: | Non-ribosomal peptide synthetase modules | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1S
 
 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant H140A/R148A in complex with C8-CoA and Leu-SNAC | | Descriptor: | Non-ribosomal peptide synthetase modules, OCTANOYL-COENZYME A, S-(2-acetamidoethyl) (2S)-2-azanyl-4-methyl-pentanethioate | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-05 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7C1K
 
 | | Crystal structure of the starter condensation domain of rhizomide synthetase RzmA mutant R148A | | Descriptor: | Non-ribosomal peptide synthetase modules | | Authors: | Zhong, L, Diao, X, Zhang, N, Li, F.W, Zhou, H.B, Chen, H.N, Ren, X, Zhang, Y, Wu, D, Bian, X. | | Deposit date: | 2020-05-04 | | Release date: | 2020-11-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Engineering and elucidation of the lipoinitiation process in nonribosomal peptide biosynthesis.
Nat Commun, 12, 2021
|
|
7V7W
 
 | | Crystal Structure of the Heterodimeric HIF-3a:ARNT Complex with oleoylethanolamide (OEA) | | Descriptor: | (Z)-N-(2-hydroxyethyl)octadec-9-enamide, Aryl hydrocarbon receptor nuclear translocator, Hypoxia-inducible factor 3-alpha | | Authors: | Diao, X, Ren, X, Li, F.W, Zhang, M, Sun, X, Wu, D. | | Deposit date: | 2021-08-21 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Identification of oleoylethanolamide as an endogenous ligand for HIF-3 alpha.
Nat Commun, 13, 2022
|
|
