8OEH
 
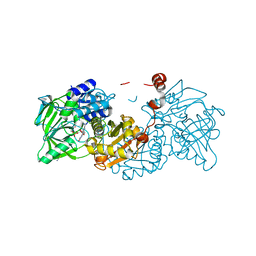 | | Aspergillus niger ferulic acid decarboxylase (Fdc) C122-S261C (DB3) variant in complex with prenylated flavin | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Roberts, G.W, Leys, D. | | Deposit date: | 2023-03-10 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Aspergillus niger ferulic acid decarboxylase (Fdc) C122-S261C (DB3) variant in complex with prenylated flavin
To Be Published
|
|
2XKR
 
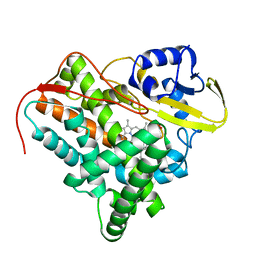 | | Crystal Structure of Mycobacterium tuberculosis CYP142: A novel cholesterol oxidase | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 142, TETRAETHYLENE GLYCOL | | Authors: | Driscoll, M, McLean, K.J, Levy, C.W, Lafite, P, Mast, N, Pikuleva, I.A, Rigby, S.E.J, Leys, D, Munro, A.W. | | Deposit date: | 2010-07-12 | | Release date: | 2010-09-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and Biochemical Characterization of Mycobacterium Tuberculosis Cyp142: Evidence for Multiple Cholesterol 27-Hydroxylase Activities in a Human Pathogen.
J.Biol.Chem., 285, 2010
|
|
8OED
 
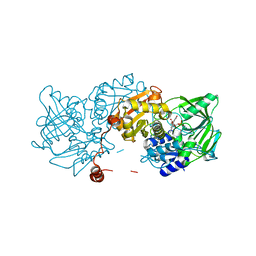 | |
5E1Z
 
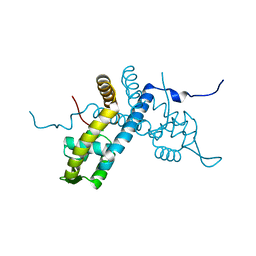 | | Crystal structure of the organohalide sensing RdhR-CbdbA1625 transcriptional regulator in the 2,4-dichlorophenol bound form | | Descriptor: | 2,4-dichlorophenol, Transcriptional regulator, MarR family | | Authors: | Quezada, C.P, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2015-09-30 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structures of RdhRCbdbA1625 provide insight into sensing of chloroaromatic compounds by MarR-type regulators
to be published
|
|
5E20
 
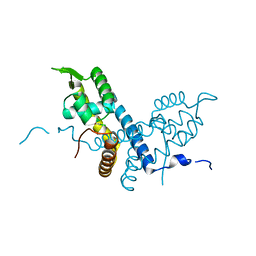 | | Crystal structure of the organohalide sensing RdhR-CbdbA1625 transcriptional regulator in the 2,3-dichlorophenol bound form | | Descriptor: | 2,3-dichlorophenol, Transcriptional regulator, MarR family | | Authors: | Quezada, C.P, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2015-09-30 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structures of RdhRCbdbA1625 provide insight into sensing of chloroaromatic compounds by MarR-type regulators
To be published
|
|
5HDI
 
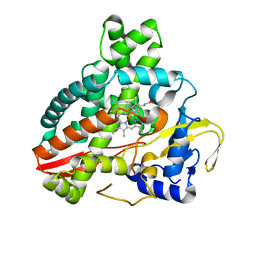 | | Structural characterization of CYP144A1, a Mycobacterium tuberculosis cytochrome P450 | | Descriptor: | Cytochrome P450 144, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chenge, J, Driscoll, M.D, McLean, K.J, Munro, A.W, Leys, D. | | Deposit date: | 2016-01-05 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural characterization of CYP144A1 - a cytochrome P450 enzyme expressed from alternative transcripts in Mycobacterium tuberculosis.
Sci Rep, 6, 2016
|
|
5E1W
 
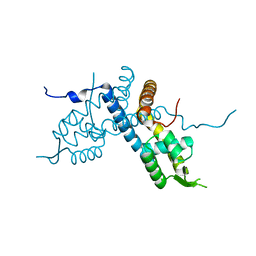 | |
5E1X
 
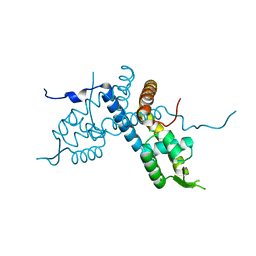 | | Crystal structure of the organohalide sensing RdhR-CbdbA1625 transcriptional regulator in the 3,4-dichlorophenol bound form | | Descriptor: | 3,4-dichlorophenol, Transcriptional regulator, MarR family | | Authors: | Quezada, C.P, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2015-09-30 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal structures of RdhRCbdbA1625 provide insight into sensing of chloroaromatic compounds by MarR-type regulators
TO BE PUBLISHED
|
|
8A9P
 
 | | Crystal structure of CYP142 from Mycobacterium tuberculosis in complex with a fragment | | Descriptor: | (3-phenyl-1,2,4-oxadiazol-5-yl)methanamine, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of CYP142 from Mycobacterium tuberculosis in complex with a fragment
To Be Published
|
|
4O1E
 
 | | Structure of a methyltransferase component in complex with MTHF involved in O-demethylation | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, Dihydropteroate synthase DHPS | | Authors: | Sjuts, H, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2013-12-15 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structures of the methyltransferase component of Desulfitobacterium hafniense DCB-2 O-demethylase shed light on methyltetrahydrofolate formation
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4O0Q
 
 | | Apo structure of a methyltransferase component involved in O-demethylation | | Descriptor: | Dihydropteroate synthase DHPS | | Authors: | Sjuts, H, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2013-12-14 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structures of the methyltransferase component of Desulfitobacterium hafniense DCB-2 O-demethylase shed light on methyltetrahydrofolate formation
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4O1F
 
 | | Structure of a methyltransferase component in complex with THF involved in O-demethylation | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, Dihydropteroate synthase DHPS | | Authors: | Sjuts, H, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2013-12-15 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the methyltransferase component of Desulfitobacterium hafniense DCB-2 O-demethylase shed light on methyltetrahydrofolate formation
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3K8D
 
 | | Crystal structure of E. coli lipopolysaccharide specific CMP-KDO synthetase in complex with CTP and 2-deoxy-Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, 3-deoxy-manno-octulosonate cytidylyltransferase, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Heyes, D.J, Levy, C.W, Lafite, P, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-10-14 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based mechanism of CMP-2-keto-3-deoxymanno-octulonic acid synthetase: convergent evolution of a sugar-activating enzyme with DNA/RNA polymerases
J.Biol.Chem., 284, 2009
|
|
2OJY
 
 | |
5M1D
 
 | | Crystal structure of N-terminally tagged UbiD from E. coli reconstituted with prFMN cofactor | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 3-octaprenyl-4-hydroxybenzoate carboxy-lyase, MANGANESE (II) ION, ... | | Authors: | Marshall, S.A, Leys, D. | | Deposit date: | 2016-10-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Oxidative Maturation and Structural Characterization of Prenylated FMN Binding by UbiD, a Decarboxylase Involved in Bacterial Ubiquinone Biosynthesis.
J. Biol. Chem., 292, 2017
|
|
3JSX
 
 | | X-ray Crystal structure of NAD(P)H: Quinone Oxidoreductase-1 (NQO1) bound to the coumarin-based inhibitor AS1 | | Descriptor: | 4-hydroxy-6,7-dimethyl-3-(naphthalen-1-ylmethyl)-2H-chromen-2-one, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Dunstan, M.S, Levy, C, Leys, D. | | Deposit date: | 2009-09-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthesis and biological evaluation of coumarin-based inhibitors of NAD(P)H: quinone oxidoreductase-1 (NQO1).
J.Med.Chem., 52, 2009
|
|
2AGZ
 
 | | Crystal structure of the carbinolamine intermediate in the reductive half-reaction of aromatic amine dehydrogenase (AADH) with tryptamine. F222 form | | Descriptor: | (1S)-1-AMINO-2-(1H-INDOL-3-YL)ETHANOL, Aromatic amine dehydrogenase, ZINC ION | | Authors: | Masgrau, L, Roujeinikova, A, Johannissen, L.O, Hothi, P, Basran, J, Ranaghan, K.E, Mulholland, A.J, Sutcliffe, M.J, Scrutton, N.S, Leys, D. | | Deposit date: | 2005-07-27 | | Release date: | 2006-04-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Atomic description of an enzyme reaction dominated by proton tunneling
Science, 312, 2006
|
|
8AA7
 
 | | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with a heme coordinated fragment | | Descriptor: | 2,3,4,9-tetrahydro-1~{H}-pyrido[3,4-b]indole, Cytochrome P450 protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-06-30 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with a heme coordinated fragment
To Be Published
|
|
2OK4
 
 | |
2OK6
 
 | |
2OIZ
 
 | |
3K8E
 
 | | Crystal structure of E. coli lipopolysaccharide specific CMP-KDO synthetase | | Descriptor: | 3-deoxy-manno-octulosonate cytidylyltransferase | | Authors: | Heyes, D.J, Levy, C.W, Lafite, P, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-10-14 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure-based mechanism of CMP-2-keto-3-deoxymanno-octulonic acid synthetase: convergent evolution of a sugar-activating enzyme with DNA/RNA polymerases
J.Biol.Chem., 284, 2009
|
|
5M0O
 
 | | Crystal structure of cytochrome P450 OleT H85Q in complex with arachidonic acid | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Tee, K.L, Munro, A, Matthews, S, Leys, D, Levy, C. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic Determinants of Alkene Production by the Cytochrome P450 Peroxygenase OleTJE.
J. Biol. Chem., 292, 2017
|
|
5M1E
 
 | | Crystal structure of N-terminally tagged UbiD from E. coli reconstituted with prFMN cofactor | | Descriptor: | (16~{R})-11,12,14,14-tetramethyl-3,5-bis(oxidanylidene)-8-[(2~{S},3~{S},4~{R})-2,3,4-tris(oxidanyl)-5-phosphonooxy-pentyl]-1,4,6,8-tetrazatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-2(7),9(17),10,12-tetraene-16-sulfonic acid, 3-octaprenyl-4-hydroxybenzoate carboxy-lyase, MANGANESE (II) ION, ... | | Authors: | Marshall, S.A, Leys, D. | | Deposit date: | 2016-10-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Oxidative Maturation and Structural Characterization of Prenylated FMN Binding by UbiD, a Decarboxylase Involved in Bacterial Ubiquinone Biosynthesis.
J. Biol. Chem., 292, 2017
|
|
5M0P
 
 | | Crystal structure of cytochrome P450 OleT F79A in complex with arachidonic acid | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, Terminal olefin-forming fatty acid decarboxylase, ... | | Authors: | Tee, K.L, Munro, A, Matthews, S, Leys, D, Levy, C. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Catalytic Determinants of Alkene Production by the Cytochrome P450 Peroxygenase OleTJE.
J. Biol. Chem., 292, 2017
|
|
