6SCB
 
 | |
2QXW
 
 | | Perdeuterated alr2 in complex with idd594 | | Descriptor: | Aldose reductase, CITRIC ACID, IDD594, ... | | Authors: | Blakeley, M.P, Ruiz, F, Cachau, R, Hazemann, I, Meilleur, F, Mitschler, A, Ginell, S, Afonine, P, Ventura, O, Cousido-Siah, A, Joachimiak, A, Myles, D, Podjarny, A. | | Deposit date: | 2007-08-13 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Quantum model of catalysis based on a mobile proton revealed by subatomic x-ray and neutron diffraction studies of h-aldose reductase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1H5P
 
 | | Solution structure of the human Sp100b SAND domain by heteronuclear NMR. | | Descriptor: | NUCLEAR AUTOANTIGEN SP100-B | | Authors: | Bottomley, M.J, Liu, Z, Collard, M.W, Huggenvik, J.I, Gibson, T.J, Sattler, M. | | Deposit date: | 2001-05-24 | | Release date: | 2001-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation.
Nat. Struct. Biol., 8, 2001
|
|
1QX4
 
 | | Structrue of S127P mutant of cytochrome b5 reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase | | Authors: | Bewley, M.C, Davis, C.A, Marohnic, C.C, Taormina, D, Barber, M.J. | | Deposit date: | 2003-09-04 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the S127P mutant of cytochrome b5 reductase that causes methemoglobinemia shows the AMP moiety of the flavin occupying the substrate binding site
Biochemistry, 42, 2003
|
|
1ZZN
 
 | |
2B58
 
 | | SSAT with coa_sp, spermine disordered, K26R mutant | | Descriptor: | COENZYME A, Diamine acetyltransferase 1 | | Authors: | Bewley, M.C, Graziano, V, Jiang, J.S, Matz, E, Studier, F.W, Pegg, A.P, Coleman, C.S, Flanagan, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-09-27 | | Release date: | 2006-01-24 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2B4D
 
 | | SSAT+COA+SP- SP disordered | | Descriptor: | COENZYME A, Diamine acetyltransferase 1 | | Authors: | Bewley, M.C, Graziano, V, Jiang, J.S, Matz, E, Studier, F.W, Pegg, A.P, Coleman, C.S, Flanagan, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-09-23 | | Release date: | 2006-01-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2B3U
 
 | | Human Spermine spermidine acetyltransferase K26R mutant | | Descriptor: | Diamine acetyltransferase 1, SULFATE ION | | Authors: | Bewley, M.C, Graziano, V, Jiang, J.S, Matz, E, Studier, F.W, Pegg, A.P, Coleman, C.S, Flanagan, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-09-21 | | Release date: | 2006-01-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2B3V
 
 | | Spermine spermidine acetyltransferase in complex with acetylcoa, K26R mutant | | Descriptor: | ACETYL COENZYME *A, Diamine acetyltransferase 1 | | Authors: | Bewley, M.C, Graziano, V, Jiang, J.S, Matz, E, Studier, F.W, Pegg, A.P, Coleman, C.S, Flanagan, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-09-21 | | Release date: | 2006-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2B4B
 
 | | SSAT+COA+BE-3-3-3, K26R mutant | | Descriptor: | COENZYME A, Diamine acetyltransferase 1, N-ETHYL-N-[3-(PROPYLAMINO)PROPYL]PROPANE-1,3-DIAMINE | | Authors: | Bewley, M.C, Graziano, V, Jiang, J.S, Matz, E, Studier, F.W, Pegg, A.P, Coleman, C.S, Flanagan, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-09-23 | | Release date: | 2006-01-17 | | Last modified: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2B5G
 
 | | Wild Type SSAT- 1.7A structure | | Descriptor: | Diamine acetyltransferase 1, SULFATE ION | | Authors: | Bewley, M.C, Graziano, V, Jiang, J.S, Matz, E, Studier, F.W, Pegg, A.P, Coleman, C.S, Flanagan, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-09-28 | | Release date: | 2006-01-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6WCY
 
 | | N160D Deamidation Mutant of Human gammaD-Crystallin | | Descriptor: | Gamma-crystallin D, SULFATE ION | | Authors: | Whitley, M.J, Rathi, N, Ambarian, M, Gronenborn, A.M. | | Deposit date: | 2020-03-31 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.204 Å) | | Cite: | Assessing the Structures and Interactions of gamma D-Crystallin Deamidation Variants.
Structure, 29, 2021
|
|
6W5B
 
 | |
8T75
 
 | |
8T74
 
 | |
5KHY
 
 | | Crystal structure of oxime-linked K6 diubiquitin | | Descriptor: | ETHANOLAMINE, Polyubiquitin-B, ZINC ION | | Authors: | Stanley, M, Virdee, S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Genetically Directed Production of Recombinant, Isosteric and Nonhydrolysable Ubiquitin Conjugates.
Chembiochem, 17, 2016
|
|
6CER
 
 | | Human pyruvate dehydrogenase complex E1 component V138M mutation | | Descriptor: | MAGNESIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, somatic form, ... | | Authors: | Whitley, M.J, Arjunan, P, Furey, W. | | Deposit date: | 2018-02-12 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Pyruvate dehydrogenase complex deficiency is linked to regulatory loop disorder in the alpha V138M variant of human pyruvate dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
6GSI
 
 | |
6GSH
 
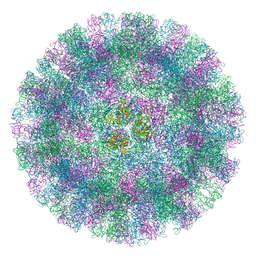 | | Feline Calicivirus Strain F9 | | Descriptor: | POTASSIUM ION, VP1 | | Authors: | Conley, M.J, Bhella, D. | | Deposit date: | 2018-06-14 | | Release date: | 2019-01-16 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Calicivirus VP2 forms a portal-like assembly following receptor engagement.
Nature, 565, 2019
|
|
5CEV
 
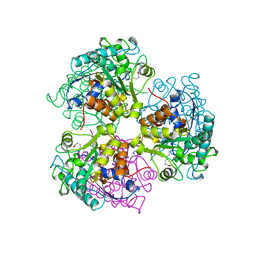 | | ARGINASE FROM BACILLUS CALDEVELOX, L-LYSINE COMPLEX | | Descriptor: | GUANIDINE, LYSINE, MANGANESE (II) ION, ... | | Authors: | Bewley, M.C, Jeffrey, P.D, Patchett, M.L, Kanyo, Z.F, Baker, E.N. | | Deposit date: | 1999-03-16 | | Release date: | 1999-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure Fold.Des., 7, 1999
|
|
4GK9
 
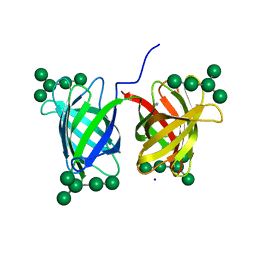 | | Crystal structure of Burkholderia oklahomensis agglutinin (BOA) bound to 3a,6a-mannopentaose | | Descriptor: | IMIDAZOLE, SODIUM ION, agglutinin (BOA), ... | | Authors: | Whitley, M.J, Furey, W, Gronenborn, A.M. | | Deposit date: | 2012-08-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Burkholderia oklahomensis agglutinin is a canonical two-domain OAA-family lectin: structures, carbohydrate binding and anti-HIV activity.
Febs J., 280, 2013
|
|
1B3E
 
 | | HUMAN SERUM TRANSFERRIN, N-TERMINAL LOBE, EXPRESSED IN PICHIA PASTORIS | | Descriptor: | CARBONATE ION, FE (III) ION, PROTEIN (SERUM TRANSFERRIN) | | Authors: | Bewley, M.C, Tam, B.M, Grewal, J, He, S, Shewry, S, Murphy, M.E.P, Mason, A.B, Woodworth, R.C, Baker, E.N, Macgillivray, R.T.A. | | Deposit date: | 1998-12-09 | | Release date: | 1999-03-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystallography and mass spectroscopy reveal that the N-lobe of human transferrin expressed in Pichia pastoris is folded correctly but is glycosylated on serine-32.
Biochemistry, 38, 1999
|
|
4GU8
 
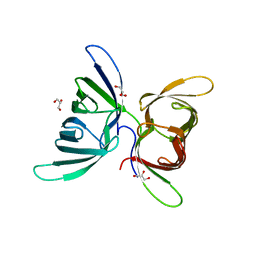 | | Crystal Structure of Burkholderia oklahomensis agglutinin (BOA) | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Burkholderia oklahomensis agglutinin (BOA), GLYCEROL | | Authors: | Whitley, M.J, Furey, W, Gronenborn, A.M. | | Deposit date: | 2012-08-29 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Burkholderia oklahomensis agglutinin is a canonical two-domain OAA-family lectin: structures, carbohydrate binding and anti-HIV activity.
Febs J., 280, 2013
|
|
2CEV
 
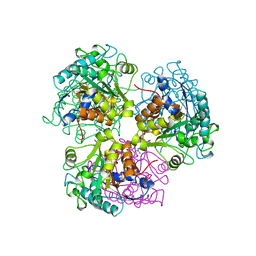 | | ARGINASE FROM BACILLUS CALDEVELOX, NATIVE STRUCTURE AT PH 8.5 | | Descriptor: | GUANIDINE, MANGANESE (II) ION, PROTEIN (ARGINASE) | | Authors: | Bewley, M.C, Jeffrey, P.D, Patchett, M.L, Kanyo, Z.F, Baker, E.N. | | Deposit date: | 1999-03-10 | | Release date: | 1999-04-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure Fold.Des., 7, 1999
|
|
5AKR
 
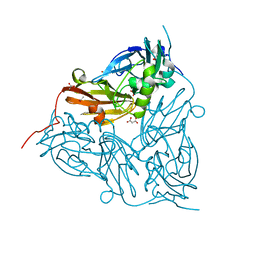 | | ATOMIC RESOLUTION STRUCTURE OF NITRITE BOUND STATE OF THE ACHROMOBACTER CYCLOCLASTES CU NITRITE REDUCTASE AT 0.87 A RESOLUTION | | Descriptor: | ACETATE ION, COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Blakeley, M.P, Hasnain, S.S, Antonyuk, S.V. | | Deposit date: | 2015-03-05 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | Sub-Atomic Resolution X-Ray Crystallography and Neutron Crystallography: Promise, Challenges and Potential.
Iucrj, 2, 2015
|
|
