8QHE
 
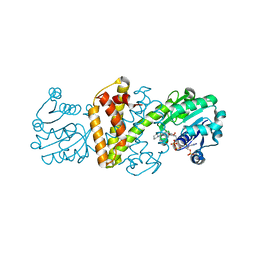 | | Crystal structure IR-09 | | Descriptor: | 6-phosphogluconate dehydrogenase NADP-binding domain-containing protein, MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Levy, C.W. | | Deposit date: | 2023-09-07 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biocatalysis in Drug Design: Engineered Reductive Aminases (RedAms) Are Used to Access Chiral Building Blocks with Multiple Stereocenters.
J.Am.Chem.Soc., 145, 2023
|
|
6FOZ
 
 | |
7A9J
 
 | |
7A9I
 
 | |
6I3C
 
 | |
6I3D
 
 | |
8PVQ
 
 | | Crystal Structure of Human PTX3 C-terminal domain | | Descriptor: | Pentraxin-related protein PTX3 | | Authors: | Levy, C.W. | | Deposit date: | 2023-07-18 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Pentraxin 3 mediated crosslinking of heavy chain hyaluronan complexes: new insights from structural and biophysical studies
To Be Published
|
|
8RA0
 
 | | Crystal structure of CysF | | Descriptor: | AMP-dependent synthetase, CITRATE ANION, TETRAETHYLENE GLYCOL | | Authors: | Levy, C.W. | | Deposit date: | 2023-11-30 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Cryptic enzymatic assembly of peptides armed with beta-lactone warheads.
Nat.Chem.Biol., 20, 2024
|
|
5CPO
 
 | | Crystal structure of XenA from Pseudomonas putida in complex with an NADH mimic (mBu) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 1-butyl-1,4,5,6-tetrahydropyridine-3-carboxamide, CALCIUM ION, ... | | Authors: | Levy, C.W. | | Deposit date: | 2015-07-21 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Better than Nature: Nicotinamide Biomimetics That Outperform Natural Coenzymes.
J.Am.Chem.Soc., 138, 2016
|
|
4KVR
 
 | | Crystal Structure of Prochlorococcus marinus aldehyde-deformylating oxygenase (mutant V41Y) | | Descriptor: | Aldehyde decarbonylase, FE (III) ION, HEXANOIC ACID | | Authors: | Levy, C.W, Khara, B, Menon, N, Mansell, D, Das, D, Marsh, E.N.G, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase.
Chembiochem, 14, 2013
|
|
4KVQ
 
 | | Crystal Structure of Prochlorococcus marinus aldehyde-deformylating oxygenase wild type with palmitic acid bound | | Descriptor: | Aldehyde decarbonylase, FE (III) ION, PALMITIC ACID | | Authors: | Levy, C.W, Khara, B, Menon, N, Mansell, D, Das, D, Marsh, E.N.G, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase.
Chembiochem, 14, 2013
|
|
1KKR
 
 | | CRYSTAL STRUCTURE OF CITROBACTER AMALONATICUS METHYLASPARTATE AMMONIA LYASE CONTAINING (2S,3S)-3-METHYLASPARTIC ACID | | Descriptor: | (2S,3S)-3-methyl-aspartic acid, 3-METHYLASPARTATE AMMONIA-LYASE, MAGNESIUM ION | | Authors: | Levy, C.W, Buckley, P.A, Sedelnikova, S, Kato, K, Asano, Y, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-12-10 | | Release date: | 2002-01-30 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights into enzyme evolution revealed by the structure of methylaspartate ammonia lyase.
Structure, 10, 2002
|
|
1KKO
 
 | | CRYSTAL STRUCTURE OF CITROBACTER AMALONATICUS METHYLASPARTATE AMMONIA LYASE | | Descriptor: | 3-METHYLASPARTATE AMMONIA-LYASE, SULFATE ION | | Authors: | Levy, C.W, Buckley, P.A, Sedelnikova, S, Kato, Y, Asano, Y, Rice, D.W, Baker, P.J. | | Deposit date: | 2001-12-10 | | Release date: | 2002-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Insights into enzyme evolution revealed by the structure of methylaspartate ammonia lyase.
Structure, 10, 2002
|
|
4KVS
 
 | | Crystal Structure of Prochlorococcus marinus aldehyde-deformylating oxygenase (mutant A134F) | | Descriptor: | Aldehyde decarbonylase, FE (III) ION, HEXANOIC ACID | | Authors: | Levy, C.W, Khara, B, Menon, N, Mansell, D, Das, D, Marsh, E.N.G, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-05-23 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase.
Chembiochem, 14, 2013
|
|
4Z43
 
 | | Crystal structure of Tryptophan 7-halogenase (PrnA) Mutant E450K | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavin-dependent tryptophan halogenase PrnA, ... | | Authors: | Levy, C.W. | | Deposit date: | 2015-04-01 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Extending the biocatalytic scope of regiocomplementary flavin-dependent halogenase enzymes.
Chem Sci, 6, 2015
|
|
5C5T
 
 | |
8BOU
 
 | | Crystal structure of Blautia producta GH94 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Levy, C.W. | | Deposit date: | 2022-11-15 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Blautia producta is a competent degrader among human gut Firmicutes for utilizing dietary beta mixed linkage glucan
To Be Published
|
|
7ZRN
 
 | |
6Z1L
 
 | |
6Z1K
 
 | |
5O4L
 
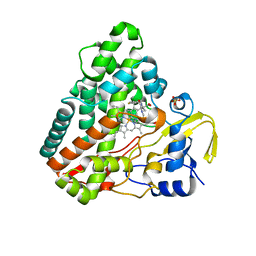 | | Crystal structure of P450 CYP121 in complex with compound 6a. | | Descriptor: | 1-[(4-fluorophenyl)methyl]-4-(3-imidazol-1-ylpropyl)piperazin-2-one, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Levy, C.W. | | Deposit date: | 2017-05-29 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Novel Aryl Substituted Pyrazoles as Small Molecule Inhibitors of Cytochrome P450 CYP121A1: Synthesis and Antimycobacterial Evaluation.
J. Med. Chem., 60, 2017
|
|
6Q7P
 
 | | Crystal structure of OE1.2 | | Descriptor: | 1,2-ETHANEDIOL, 1-PHENYLETHANONE, MAGNESIUM ION, ... | | Authors: | Levy, C.W. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Design and evolution of an enzyme with a non-canonical organocatalytic mechanism.
Nature, 570, 2019
|
|
6Q7R
 
 | |
5O4K
 
 | | Crystal structure of P450 CYP121 in complex with compound 6b. | | Descriptor: | 1-[(4-chlorophenyl)methyl]-4-(3-imidazol-1-ylpropyl)piperazin-2-one, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Levy, C.W. | | Deposit date: | 2017-05-29 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel Aryl Substituted Pyrazoles as Small Molecule Inhibitors of Cytochrome P450 CYP121A1: Synthesis and Antimycobacterial Evaluation.
J. Med. Chem., 60, 2017
|
|
6Q7N
 
 | |
