2BTL
 
 | |
2C9K
 
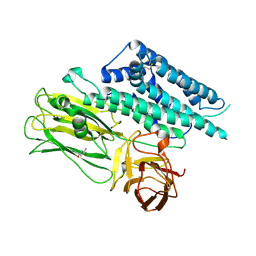 | | Structure of the functional form of the mosquito-larvicidal Cry4Aa toxin from Bacillus thuringiensis at 2.8 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PESTICIDAL CRYSTAL PROTEIN CRY4AA | | Authors: | Boonserm, P, Mo, M, Angsuthanasombat, C, Lescar, J. | | Deposit date: | 2005-12-13 | | Release date: | 2006-04-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Functional Form of the Mosquito Larvicidal Cry4Aa Toxin from Bacillus Thuringiensis at 2.8-Angstrom Resolution.
J.Bacteriol., 188, 2006
|
|
7DIJ
 
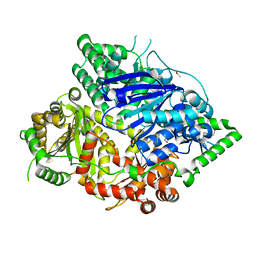 | | Falcilysin in complex with MK-4815 | | Descriptor: | 1,2-ETHANEDIOL, 2-(aminomethyl)-3,5-ditert-butyl-phenol, ACETATE ION, ... | | Authors: | Lin, J.Q, El Sahili, A, Lescar, J. | | Deposit date: | 2020-11-19 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protease in complex with inhibitor
To Be Published
|
|
7DI7
 
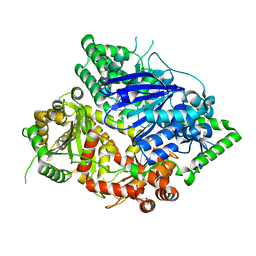 | | Falcilysin in complex with chloroquine | | Descriptor: | ACETATE ION, Falcilysin, N4-(7-CHLORO-QUINOLIN-4-YL)-N1,N1-DIETHYL-PENTANE-1,4-DIAMINE, ... | | Authors: | Lin, J.Q, El Sahili, A, Lescar, J. | | Deposit date: | 2020-11-18 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Protease in complex with compound
To Be Published
|
|
7DU3
 
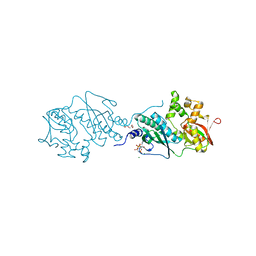 | | ThiL in complex with AMP-PNP | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Lin, J.Q, Lescar, J. | | Deposit date: | 2021-01-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | ThiL in complex with AMP-PNP
To Be Published
|
|
7F8C
 
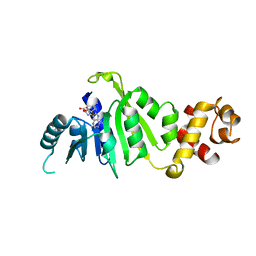 | |
7F8A
 
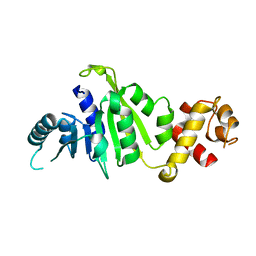 | | Crystal structure of rRNA methyltransferase Erm38 | | Descriptor: | Erm(38) | | Authors: | Goh, B.C, Lescar, J. | | Deposit date: | 2021-07-01 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of mycobacterial erythromycin resistance methyltransferase Erm38 reveals its RNA-binding site.
J.Biol.Chem., 298, 2022
|
|
7F8B
 
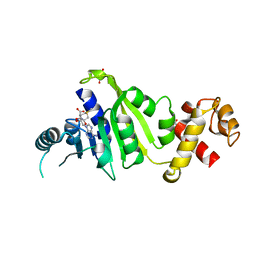 | |
7F5Q
 
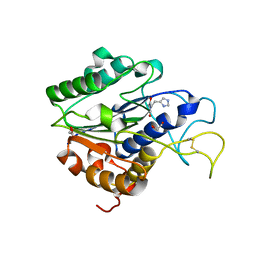 | | The crystal structure of VyPAL2 peptide asparaginyl ligase in its active enzyme form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, S, Sahili, A, Lescar, J. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.
Plant Cell, 34, 2022
|
|
7F5J
 
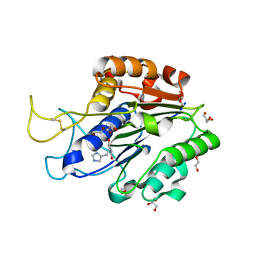 | | The crystal structure of VyPAL2-I244V, a more efficient mutant of VyPAL2 peptide asparaginyl ligase in its active enzyme form | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, S, Sahili, A, Lescar, J. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.593 Å) | | Cite: | Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.
Plant Cell, 34, 2022
|
|
7F5P
 
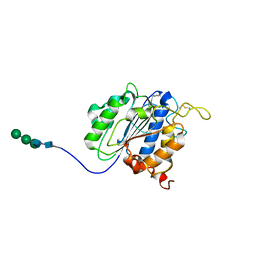 | | The crystal structure of VyPAL2-C214A, a dead mutant of VyPAL2 peptide asparaginyl ligase in form I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Peptide Asparaginyl Ligases, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, S, Sahili, A, Lescar, J. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.
Plant Cell, 34, 2022
|
|
7FA0
 
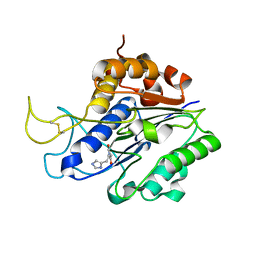 | | The crystal structure of VyPAL2-C214A, a dead mutant of VyPAL2 peptide asparaginyl ligase in form II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Peptide Asparaginyl Ligases, ... | | Authors: | Hu, S, Sahili, A, Lescar, J. | | Deposit date: | 2021-07-05 | | Release date: | 2022-07-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.
Plant Cell, 34, 2022
|
|
5WYR
 
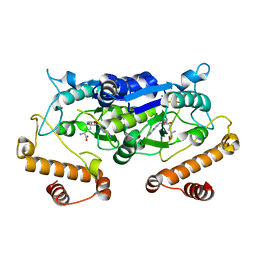 | | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | SINEFUNGIN, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W.H, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2017-01-15 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 2019
|
|
2J7W
 
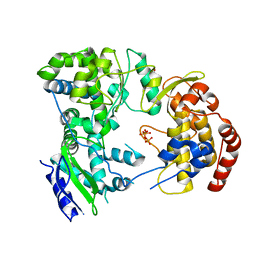 | | Dengue virus NS5 RNA dependent RNA polymerase domain complexed with 3' dGTP | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-TRIPHOSPHATE, POLYPROTEIN, ... | | Authors: | Yap, T.L, Xu, T, Chen, Y.L, Malet, H, Egloff, M.P, Canard, B, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Dengue Virus RNA- Dependent RNA Polymerase Catalytic Domain at 1.85 Angstrom Resolution.
J.Virol., 81, 2007
|
|
2J7U
 
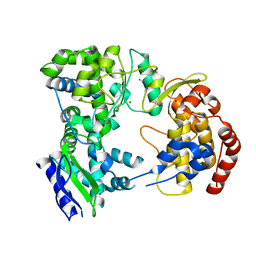 | | Dengue virus NS5 RNA dependent RNA polymerase domain | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, RNA DEPENDENT RNA POLYMERASE, ... | | Authors: | Yap, T.L, Xu, T, Chen, Y.L, Malet, H, Egloff, M.P, Canard, B, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Dengue Virus RNA- Dependent RNA Polymerase Catalytic Domain at 1.85 Angstrom Resolution.
J.Virol., 81, 2007
|
|
5ZQK
 
 | | Dengue Virus Non Structural Protein 5 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | El Sahili, A, Soh, T.S, Schiltz, J, Gharbi-Ayachi, A, Goh, B.C, Seh, C.C, Dedon, P.C, Shi, P.Y, Lim, S.P, Lescar, J. | | Deposit date: | 2018-04-19 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NS5 from Dengue Virus Serotype 2 Can Adopt a Conformation Analogous to That of Its Zika Virus and Japanese Encephalitis Virus Homologues.
J.Virol., 94, 2019
|
|
6AFK
 
 | | Crystal structure of TrmD from Pseudomonas aeruginosa in complex with active-site inhibitor | | Descriptor: | N-{(3S)-1-[3-(pyridin-4-yl)-1H-pyrazol-5-yl]piperidin-3-yl}-1H-indole-2-carboxamide, S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Koay, A, Wong, Y.W, Sahili, A.E, Nah, Q, Kang, C, Poulsen, A, Chionh, Y.K, McBee, M, Matter, A, Hill, J, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Targeting the Bacterial Epitranscriptome for Antibiotic Development: Discovery of Novel tRNA-(N1G37) Methyltransferase (TrmD) Inhibitors.
Acs Infect Dis., 5, 2019
|
|
2M1C
 
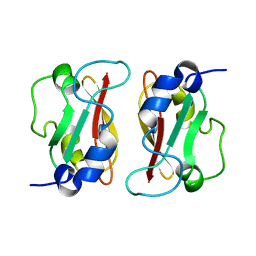 | | HADDOCK structure of GtYybT PAS Homodimer | | Descriptor: | DHH subfamily 1 protein | | Authors: | Liang, Z.X, Pervushin, K, Tan, E, Rao, F, Pasunooti, S, Soehano, I, Lescar, J. | | Deposit date: | 2012-11-25 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the PAS Domain of a Thermophilic YybT Protein Homolog Reveals a Potential Ligand-binding Site.
J.Biol.Chem., 288, 2013
|
|
5XGB
 
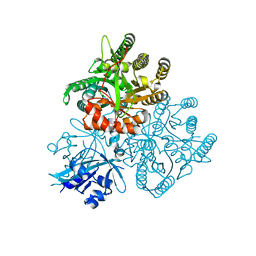 | |
5XAJ
 
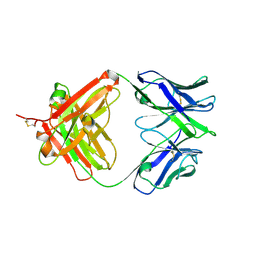 | |
5XGE
 
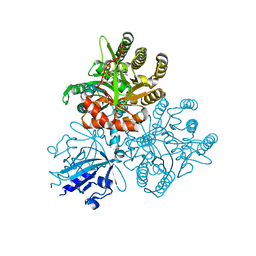 | | Crystal structure of the PAS-GGDEF-EAL domain of PA0861 from Pseudomonas aeruginosa in complex with cyclic di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Uncharacterized protein PA0861 | | Authors: | Liu, C, Liew, C.W, Sreekanth, R, Lescar, J. | | Deposit date: | 2017-04-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Insights into Biofilm Dispersal Regulation from the Crystal Structure of the PAS-GGDEF-EAL Region of RbdA from Pseudomonas aeruginosa.
J. Bacteriol., 200, 2018
|
|
5XGD
 
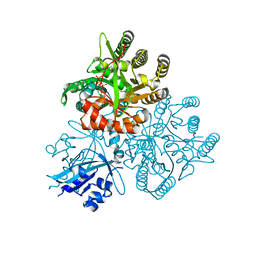 | | Crystal structure of the PAS-GGDEF-EAL domain of PA0861 from Pseudomonas aeruginosa in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Uncharacterized protein PA0861 | | Authors: | Liu, C, Liew, C.W, Sreekanth, R, Lescar, J. | | Deposit date: | 2017-04-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into Biofilm Dispersal Regulation from the Crystal Structure of the PAS-GGDEF-EAL Region of RbdA from Pseudomonas aeruginosa.
J. Bacteriol., 200, 2018
|
|
5Y41
 
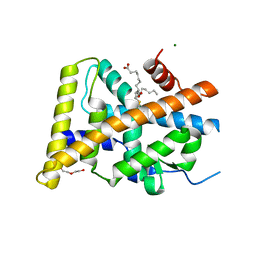 | | Crystal Structure of LIGAND-BOUND NURR1-LBD | | Descriptor: | (13E,15S)-15-hydroxy-9-oxoprosta-10,13-dien-1-oic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sreekanth, R, Lescar, J, Yoon, H.S. | | Deposit date: | 2017-07-31 | | Release date: | 2018-12-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | PGE1 and PGA1 bind to Nurr1 and activate its transcriptional function.
Nat.Chem.Biol., 2020
|
|
5WSA
 
 | | Pyruvate kinase (PYK) from Mycobacterium tuberculosis in complex with Oxalate and allosteric activator Glucose 6-Phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Zhong, W, Cai, Q, El Sahili, A, Lescar, J, Dedon, P.C. | | Deposit date: | 2016-12-06 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Allosteric pyruvate kinase-based "logic gate" synergistically senses energy and sugar levels in Mycobacterium tuberculosis.
Nat Commun, 8, 2017
|
|
5WSC
 
 | | Crystal of pyruvate kinase (PYK) from Mycobacterium tuberculosis in complex with Oxalate, soaked with allosteric activators AMP and Glucose 6-Phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhong, W, Cai, Q, El Sahili, A, Lescar, J, Dedon, P.C. | | Deposit date: | 2016-12-06 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric pyruvate kinase-based "logic gate" synergistically senses energy and sugar levels in Mycobacterium tuberculosis.
Nat Commun, 8, 2017
|
|
