6Y2M
 
 | | Streptavidin mutant S112R with a biotC4-1 cofactor - an artificial iron hydroxylase | | Descriptor: | Streptavidin, biotC4-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6Y2T
 
 | | Streptavidin wildtype with a biotC4-1 cofactor - an artificial iron hydroxylase | | Descriptor: | GLYCEROL, Streptavidin, biotC4-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6Y3Q
 
 | | Streptavidin mutant S112R_K121E with a biotC5-1 cofactor - an artificial iron hydroxylase | | Descriptor: | SULFATE ION, Streptavidin, biotC5-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-18 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6Y25
 
 | | Streptavidin mutant S112R,K121E with a biotC4-1 cofactor - an artificial iron hydroxylase | | Descriptor: | Streptavidin, biotC4-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-14 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
6Y34
 
 | | Streptavidin wildtype with a biotC5-1 cofactor - an artificial iron hydroxylase | | Descriptor: | GLYCEROL, Streptavidin, biotC5-1 cofactor | | Authors: | Serrano-Plana, J, Rumo, C, Rebelein, J.G, Peterson, R.L, Barnet, M, Ward, T.R. | | Deposit date: | 2020-02-17 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.307 Å) | | Cite: | Enantioselective Hydroxylation of Benzylic C(sp3)-H Bonds by an Artificial Iron Hydroxylase Based on the Biotin-Streptavidin Technology.
J.Am.Chem.Soc., 142, 2020
|
|
8PBB
 
 | | CHAPSO treated partial catalytic component (comprising only AnfD & AnfK, lacking AnfG and FeFeco) of iron nitrogenase from Rhodobacter capsulatus | | Descriptor: | FE(8)-S(7) CLUSTER, Nitrogenase iron-iron protein, beta subunit, ... | | Authors: | Schmidt, F.V, Schulz, L, Zarzycki, J, Prinz, S, Erb, T.J, Rebelein, J.G. | | Deposit date: | 2023-06-09 | | Release date: | 2023-10-04 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structural insights into the iron nitrogenase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6QN0
 
 | |
6QNL
 
 | |
7YZZ
 
 | | Crystal structure of Vibrio alkaline phosphatase in 0.5 M NaCl | | Descriptor: | 1,2-ETHANEDIOL, Alkaline phosphatase, CHLORIDE ION, ... | | Authors: | Markusson, S, Hjorleifsson, J.G, Kursula, P, Asgeirsson, B. | | Deposit date: | 2022-02-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structural Characterization of Functionally Important Chloride Binding Sites in the Marine Vibrio Alkaline Phosphatase.
Biochemistry, 61, 2022
|
|
6QNG
 
 | |
4YA8
 
 | | structure of plasmepsin II from Plasmodium Falciparum complexed with inhibitor PG394 | | Descriptor: | GLYCEROL, N'-[(2S,3S)-3-hydroxy-1-phenyl-4-{[2-(pyridin-2-yl)propan-2-yl]amino}butan-2-yl]-N,N-dipropyl-5-(pyridin-1(2H)-yl)benzene-1,3-dicarboxamide, Plasmepsin-2 | | Authors: | Recacha, R, Leitans, J, Tars, K, Jaudzems, K. | | Deposit date: | 2015-02-17 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Structures of plasmepsin II from Plasmodium falciparum in complex with two hydroxyethylamine-based inhibitors.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
7Z00
 
 | | Crystal structure of Vibrio alkaline phosphatase in 1.0 M KBr | | Descriptor: | Alkaline phosphatase, BROMIDE ION, MAGNESIUM ION, ... | | Authors: | Markusson, S, Hjorleifsson, J.G, Kursula, P, Asgeirsson, B. | | Deposit date: | 2022-02-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Characterization of Functionally Important Chloride Binding Sites in the Marine Vibrio Alkaline Phosphatase.
Biochemistry, 61, 2022
|
|
1B4Y
 
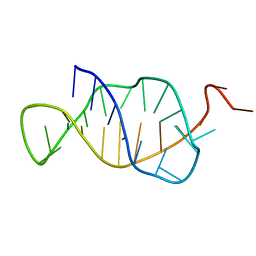 | | STRUCTURE AND MECHANISM OF FORMATION OF THE H-Y5 ISOMER OF AN INTRAMOLECULAR DNA TRIPLE HELIX. | | Descriptor: | DNA (H-Y5 TRIPLE HELIX) | | Authors: | Van Dongen, M.J.P, Doreleijers, J.F, Van Der Marel, G.A, Van Boom, J.H, Hilbers, C.W, Wijmenga, S.S. | | Deposit date: | 1998-12-30 | | Release date: | 1999-09-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and mechanism of formation of the H-y5 isomer of an intramolecular DNA triple helix.
Nat.Struct.Biol., 6, 1999
|
|
1AW9
 
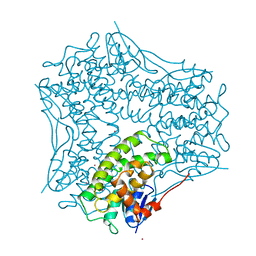 | | STRUCTURE OF GLUTATHIONE S-TRANSFERASE III IN APO FORM | | Descriptor: | CADMIUM ION, GLUTATHIONE S-TRANSFERASE III | | Authors: | Neuefeind, T, Huber, R, Reinemer, P, Knaeblein, J. | | Deposit date: | 1997-10-13 | | Release date: | 1998-10-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cloning, sequencing, crystallization and X-ray structure of glutathione S-transferase-III from Zea mays var. mutin: a leading enzyme in detoxification of maize herbicides.
J.Mol.Biol., 274, 1997
|
|
4Z22
 
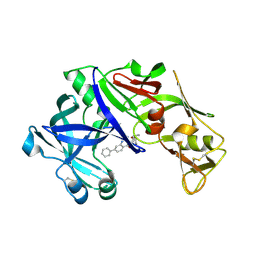 | | structure of plasmepsin II from Plasmodium Falciparum complexed with inhibitor DR718A | | Descriptor: | 2-amino-7-phenyl-3-{[(2R,5S)-5-phenyltetrahydrofuran-2-yl]methyl}quinazolin-4(3H)-one, Plasmepsin-2 | | Authors: | Recacha, R, Leitans, J, Tars, K, Jaudzems, K. | | Deposit date: | 2015-03-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Fragment-Based Discovery of 2-Aminoquinazolin-4(3H)-ones As Novel Class Nonpeptidomimetic Inhibitors of the Plasmepsins I, II, and IV.
J.Med.Chem., 59, 2016
|
|
5CW9
 
 | | Crystal structure of De novo designed ferredoxin-ferredoxin domain insertion protein | | Descriptor: | De novo designed ferredoxin-ferredoxin domain insertion protein | | Authors: | DiMaio, F, King, I.C, Gleixner, J, Doyle, L, Stoddard, B, Baker, D. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.108 Å) | | Cite: | Precise assembly of complex beta sheet topologies from de novo designed building blocks.
Elife, 4, 2015
|
|
4PCB
 
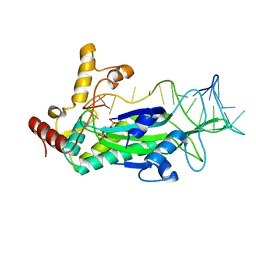 | | Conjugative Relaxase TrwC in complex with mutant OriT Dna | | Descriptor: | DNA 5'-D(P*GP*CP*AP*CP*CP*GP*AP*AP*GP*GP*TP*GP*CP*GP*TP*AP*TP*TP*CP*TP*TP*GP - 3'), PHOSPHATE ION, TrwC | | Authors: | Moncalian, G, Carballeira, J.D, de la Cruz, F, Gonzalez-Perez, B. | | Deposit date: | 2014-04-14 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A high security double lock and key mechanism in HUH relaxases controls oriT-processing for plasmid conjugation.
Nucleic Acids Res., 42, 2014
|
|
2OPR
 
 | | Crystal Structure of K101E Mutant HIV-1 Reverse Transcriptase in Complex with GW420867X. | | Descriptor: | ISOPROPYL (2S)-2-ETHYL-7-FLUORO-3-OXO-3,4-DIHYDROQUINOXALINE-1(2H)-CARBOXYLATE, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Weaver, K.L, Short, S.A, Chan, J.H, Kleim, J, Stammers, D.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Relationship of Potency and Resilience to Drug Resistant Mutations for GW420867X Revealed by Crystal Structures of Inhibitor Complexes for Wild-Type, Leu100Ile, Lys101Glu, and Tyr188Cys Mutant HIV-1 Reverse Transcriptases.
J.Med.Chem., 50, 2007
|
|
2OPS
 
 | | Crystal Structure of Y188C Mutant HIV-1 Reverse Transcriptase in Complex with GW420867X. | | Descriptor: | ISOPROPYL (2S)-2-ETHYL-7-FLUORO-3-OXO-3,4-DIHYDROQUINOXALINE-1(2H)-CARBOXYLATE, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Weaver, K.L, Short, S.A, Chan, J.H, Kleim, J, Stammers, D.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Relationship of Potency and Resilience to Drug Resistant Mutations for GW420867X Revealed by Crystal Structures of Inhibitor Complexes for Wild-Type, Leu100Ile, Lys101Glu, and Tyr188Cys Mutant HIV-1 Reverse Transcriptases.
J.Med.Chem., 50, 2007
|
|
1Z2M
 
 | | Crystal Structure of ISG15, the Interferon-Induced Ubiquitin Cross Reactive Protein | | Descriptor: | OSMIUM 4+ ION, interferon, alpha-inducible protein (clone IFI-15K) | | Authors: | Narasimhan, J, Wang, M, Fu, Z, Klein, J.M, Haas, A.L, Kim, J.J. | | Deposit date: | 2005-03-08 | | Release date: | 2005-05-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Interferon-induced Ubiquitin-like Protein ISG15.
J.Biol.Chem., 280, 2005
|
|
2OPQ
 
 | | Crystal Structure of L100I Mutant HIV-1 Reverse Transcriptase in Complex with GW420867X. | | Descriptor: | ISOPROPYL (2S)-2-ETHYL-7-FLUORO-3-OXO-3,4-DIHYDROQUINOXALINE-1(2H)-CARBOXYLATE, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Weaver, K.L, Short, S.A, Chan, J.H, Kleim, J, Stammers, D.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Relationship of Potency and Resilience to Drug Resistant Mutations for GW420867X Revealed by Crystal Structures of Inhibitor Complexes for Wild-Type, Leu100Ile, Lys101Glu, and Tyr188Cys Mutant HIV-1 Reverse Transcriptases.
J.Med.Chem., 50, 2007
|
|
2OPP
 
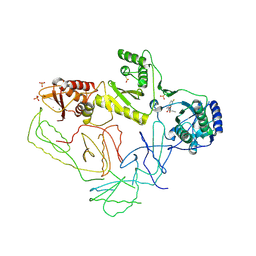 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with GW420867X. | | Descriptor: | ISOPROPYL (2S)-2-ETHYL-7-FLUORO-3-OXO-3,4-DIHYDROQUINOXALINE-1(2H)-CARBOXYLATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Weaver, K.L, Chan, S.J.H, Kleim, J, Stammers, D.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Relationship of Potency and Resilience to Drug Resistant Mutations for GW420867X Revealed by Crystal Structures of Inhibitor Complexes for Wild-Type, Leu100Ile, Lys101Glu, and Tyr188Cys Mutant HIV-1 Reverse Transcriptases.
J.Med.Chem., 50, 2007
|
|
1LRI
 
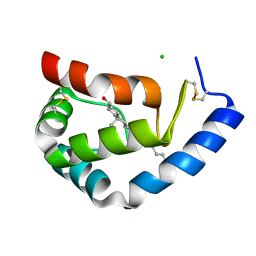 | | BETA-CRYPTOGEIN-CHOLESTEROL COMPLEX | | Descriptor: | Beta-elicitin cryptogein, CHLORIDE ION, CHOLESTEROL | | Authors: | Lascombe, M.-B, Ponchet, M, Venard, P, Milat, M.-L, Blein, J.-P, Prange, T. | | Deposit date: | 2002-05-15 | | Release date: | 2002-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The 1.45 A resolution structure of the cryptogein-cholesterol complex: a close-up view of a sterol carrier protein (SCP) active site.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4KYZ
 
 | | Three-dimensional structure of triclinic form of de novo design insertion domain, Northeast Structural Genomics Consortium (NESG) Target OR327 | | Descriptor: | Designed protein OR327 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Gleixner, J, Baker, D, Everett, J.K, Acton, T.B, Kornhaber, G, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Precise assembly of complex beta sheet topologies from de novo designed building blocks.
Elife, 4, 2015
|
|
4KY3
 
 | | Three-dimensional Structure of the orthorhombic crystal of computationally designed insertion domain , Northeast Structural Genomics Consortium (NESG) Target OR327 | | Descriptor: | designed protein OR327 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Gleixner, J, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-28 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.964 Å) | | Cite: | Precise assembly of complex beta sheet topologies from de novo designed building blocks.
Elife, 4, 2015
|
|
