8E1X
 
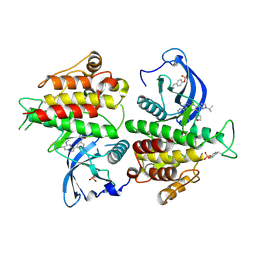 | | FGFR2 kinase domain in complex with a Pyrazolo[1,5-a]pyrimidine analog (Compound 29) | | Descriptor: | (5M)-N-methyl-5-{(6M,8S)-5-{[(3S)-oxolan-3-yl]amino}-6-[1-(propan-2-yl)-1H-pyrazol-3-yl]pyrazolo[1,5-a]pyrimidin-3-yl}pyridine-3-carboxamide, Fibroblast growth factor receptor 2 | | Authors: | Lei, H.-T, Epling, L.B, Deller, M.C. | | Deposit date: | 2022-08-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Wild-Type and Gatekeeper Mutant Fibroblast Growth Factor Receptor (FGFR) 2/3.
J.Med.Chem., 65, 2022
|
|
6XNK
 
 | |
7KGV
 
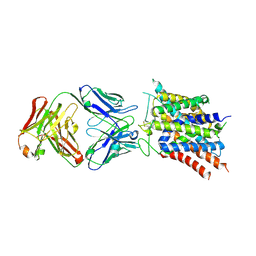 | | Crystal structure of sodium-coupled neutral amino acid transporter SLC38A9 in the N-terminal plugged form | | Descriptor: | Monoclonal antibody Fab heavy chain, Monoclonal antibody Fab light chain, Sodium-coupled neutral amino acid transporter 9 | | Authors: | Lei, H, Mu, X, Hattne, J, Gonen, T. | | Deposit date: | 2020-10-19 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A conformational change in the N terminus of SLC38A9 signals mTORC1 activation.
Structure, 29, 2021
|
|
4RNA
 
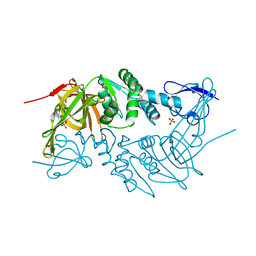 | | Crystal structure of PLpro from Middle East Respiratory Syndrome (MERS) coronavirus | | Descriptor: | PHOSPHATE ION, ZINC ION, papain-like protease | | Authors: | Lei, H, Santarsiero, B.D, Lee, H, Johnson, M.E. | | Deposit date: | 2014-10-23 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | Inhibitor Recognition Specificity of MERS-CoV Papain-like Protease May Differ from That of SARS-CoV.
Acs Chem.Biol., 10, 2015
|
|
4YIW
 
 | | DIHYDROOROTASE FROM BACILLUS ANTHRACIS WITH SUBSTRATE BOUND | | Descriptor: | Dihydroorotase, N-CARBAMOYL-L-ASPARTATE, ZINC ION | | Authors: | Lei, H, Santarsiero, B.D, Rice, A.J, Lee, H, Johnson, M.E. | | Deposit date: | 2015-03-02 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Ca-asp bound X-ray structure and inhibition of Bacillus anthracis dihydroorotase (DHOase).
Bioorg.Med.Chem., 24, 2016
|
|
7MRI
 
 | | Crystal structure of N63T yeast iso-1-cytochrome c | | Descriptor: | Cytochrome c isoform 1, HEME C | | Authors: | Lei, H, Bowler, B.E, Evenson, G.E. | | Deposit date: | 2021-05-07 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Effect on intrinsic peroxidase activity of substituting coevolved residues from Omega-loop C of human cytochrome c into yeast iso-1-cytochrome c.
J.Inorg.Biochem., 232, 2022
|
|
6C08
 
 | | Zebrafish SLC38A9 with arginine bound in the cytosol open state | | Descriptor: | ARGININE, Sodium-coupled neutral amino acid transporter 9, antibody Fab Heavy Chain, ... | | Authors: | Lei, H.-T, Gonen, T. | | Deposit date: | 2017-12-28 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Crystal structure of arginine-bound lysosomal transporter SLC38A9 in the cytosol-open state.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4PT5
 
 | |
3QPS
 
 | | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni | | Descriptor: | CHOLIC ACID, CmeR | | Authors: | Lei, H.T, Routh, M.D, Shen, Z, Su, C.C, Zhang, Q, Yu, E.W. | | Deposit date: | 2011-02-14 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni.
Protein Sci., 20, 2011
|
|
3QQA
 
 | | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni | | Descriptor: | CmeR, TAUROCHOLIC ACID | | Authors: | Lei, H.T, Routh, M.D, Shen, Z, Su, C.-C, Zhang, Q, Yu, E.W. | | Deposit date: | 2011-02-15 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of CmeR-bile acid complexes from Campylobacter jejuni.
Protein Sci., 20, 2011
|
|
6DUJ
 
 | | Crystal structure of A51V variant of Human Cytochrome c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Lei, H, Bowler, B.E. | | Deposit date: | 2018-06-20 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82202721 Å) | | Cite: | Naturally Occurring A51V Variant of Human CytochromecDestabilizes the Native State and Enhances Peroxidase Activity.
J.Phys.Chem.B, 123, 2019
|
|
3SWW
 
 | | Crystal structure of human dpp-iv in complex with sa-(+)-3-(aminomethyl)-4-(2,4-dichlorophenyl)-6-(2-methoxyphenyl)- 2-methyl-5h-pyrrolo[3,4-b]pyridin-7(6h)-one | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(aminomethyl)-4-(2,4-dichlorophenyl)-6-(2-methoxyethyl)-2-methyl-5,6-dihydro-7H-pyrrolo[3,4-b]pyridin-7-one, ... | | Authors: | Klei, H.E. | | Deposit date: | 2011-07-14 | | Release date: | 2011-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 7-Oxopyrrolopyridine-derived DPP4 inhibitors-mitigation of CYP and hERG liabilities via introduction of polar functionalities in the active site.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SW2
 
 | | X-ray crystal structure of human FXA in complex with 6-chloro-N-((3S)-2-oxo-1-(2-oxo-2-((5S)-8-oxo-5,6-dihydro-1H-1,5-methanopyrido[1,2-a][1,5]diazocin-3(2H,4H,8H)-yl)ethyl)piperidin-3-yl)naphthalene-2-sulfonamide | | Descriptor: | 6-chloro-N-((3S)-2-oxo-1-(2-oxo-2-((5S)-8-oxo-5,6-dihydro-1H-1,5-methanopyrido[1,2-a][1,5]diazocin-3(2H,4H,8H)-yl)ethyl)piperidin-3-yl)naphthalene-2-sulfonamide, CALCIUM ION, Coagulation factor X, ... | | Authors: | Klei, H.E. | | Deposit date: | 2011-07-13 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Arylsulfonamidopiperidone derivatives as a novel class of factor Xa inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1HJ8
 
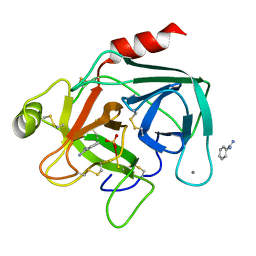 | | 1.00 AA Trypsin from Atlantic Salmon | | Descriptor: | BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Leiros, H.-K.S, Mcsweeney, S.M, Smalas, A.O. | | Deposit date: | 2001-01-09 | | Release date: | 2002-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic Resolution Structure of Trypsin Provide Insight Into Structural Radiation Damage
Acta Crystallogr.,Sect.D, 57, 2001
|
|
2FXD
 
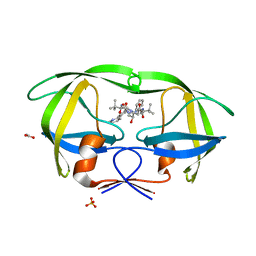 | | X-ray crystal structure of HIV-1 protease IRM mutant complexed with atazanavir (BMS-232632) | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, ACETATE ION, ... | | Authors: | Klei, H.E, Sheriff, S. | | Deposit date: | 2006-02-04 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystal structures of human immunodeficiency virus type 1 protease mutants complexed with atazanavir.
J.Virol., 81, 2007
|
|
1W3P
 
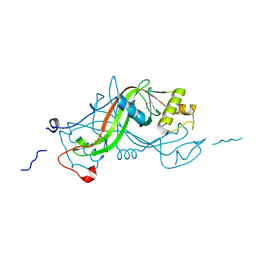 | | NimA from D. radiodurans with a His71-Pyruvate residue | | Descriptor: | ACETATE ION, NIMA-RELATED PROTEIN, PYRUVIC ACID | | Authors: | Leiros, H.-K.S, Kozielski-Stuhrmann, S, Kapp, U, Terradot, L, Leonard, G.A, McSweeney, S.M. | | Deposit date: | 2004-07-17 | | Release date: | 2004-10-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of 5-Nitroimidazole Antibiotic Resistance: The Crystal Structure of Nima from Deinococcus Radiodurans
J.Biol.Chem., 279, 2004
|
|
1W3R
 
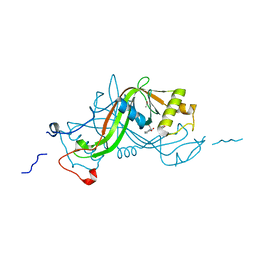 | | NimA from D. radiodurans with Metronidazole and Pyruvate | | Descriptor: | ACETATE ION, Metronidazole, NIMA-RELATED PROTEIN, ... | | Authors: | Leiros, H.-K.S, Kozielski-Stuhrmann, S, Kapp, U, Terradot, L, Leonard, G.A, McSweeney, S.M. | | Deposit date: | 2004-07-17 | | Release date: | 2004-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of 5-Nitroimidazole Antibiotic Resistance: The Crystal Structure of Nima from Deinococcus Radiodurans
J.Biol.Chem., 279, 2004
|
|
1W3O
 
 | | Crystal structure of NimA from D. radiodurans | | Descriptor: | ACETATE ION, NIMA-RELATED PROTEIN, PYRUVIC ACID | | Authors: | Leiros, H.-K.S, Kozielski-Stuhrmann, S, Kapp, U, Terradot, L, Leonard, G.A, McSweeney, S.M. | | Deposit date: | 2004-07-17 | | Release date: | 2004-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of 5-Nitroimidazole Antibiotic Resistance: The Crystal Structure of Nima from Deinococcus Radiodurans
J.Biol.Chem., 279, 2004
|
|
1W3Q
 
 | | NimA from D. radiodurans with covalenly bound lactate | | Descriptor: | ACETATE ION, LACTIC ACID, NIMA-RELATED PROTEIN | | Authors: | Leiros, H.-K.S, Kozielski-Stuhrmann, S, Kapp, U, Terradot, L, Leonard, G.A, Mcsweeney, S.M. | | Deposit date: | 2004-07-17 | | Release date: | 2004-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Basis of 5-Nitroimidazole Antibiotic Resistance: The Crystal Structure of Nima from Deinococcus Radiodurans
J.Biol.Chem., 279, 2004
|
|
7OBN
 
 | | Structural investigations of a new L3 DNA ligase: structure-function analysis | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3'), DNA ligase, ... | | Authors: | Leiros, H.-K.S, Williamson, A. | | Deposit date: | 2021-04-23 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Bacteriophage origin of some minimal ATP-dependent DNA ligases: a new structure from Burkholderia pseudomallei with striking similarity to Chlorella virus ligase.
Sci Rep, 11, 2021
|
|
1UTN
 
 | | Trypsin specificity as elucidated by LIE calculations, X-ray structures and association constant measurements | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BENZYLAMINE, CALCIUM ION, ... | | Authors: | Leiros, H.-K.S, Brandsdal, B.O, Andersen, O.A, Os, V, Leiros, I, Helland, R, Otlewski, J, Willassen, N.P, Smalas, A.O. | | Deposit date: | 2003-12-09 | | Release date: | 2004-01-09 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Trypsin Specificity as Elucidated by Lie Calculations, X-Ray Structures, and Association Constant Measurements
Protein Sci., 13, 2004
|
|
1UTP
 
 | | Trypsin specificity as elucidated by LIE calculations, X-ray structures and association constant measurements | | Descriptor: | 4-PHENYLBUTYLAMINE, CALCIUM ION, GLYCEROL, ... | | Authors: | Leiros, H.-K.S, Brandsdal, B.O, Andersen, O.A, Os, V, Leiros, I, Helland, R, Otlewski, J, Willassen, N.P, Smalas, A.O. | | Deposit date: | 2003-12-09 | | Release date: | 2004-01-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Trypsin Specificity as Elucidated by Lie Calculations, X-Ray Structures, and Association Constant Measurements
Protein Sci., 13, 2004
|
|
1UTQ
 
 | | Trypsin specificity as elucidated by LIE calculations, X-ray structures and association constant measurements | | Descriptor: | CALCIUM ION, GLYCEROL, TRYPSINOGEN | | Authors: | Leiros, H.-K.S, Brandsdal, B.O, Andersen, O.A, Os, V, Leiros, I, Helland, R, Otlewski, J, Willassen, N.P, Smalas, A.O. | | Deposit date: | 2003-12-09 | | Release date: | 2004-01-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Trypsin Specificity as Elucidated by Lie Calculations, X-Ray Structures, and Association Constant Measurements
Protein Sci., 13, 2004
|
|
1UTO
 
 | | Trypsin specificity as elucidated by LIE calculations, X-ray structures and association constant measurements | | Descriptor: | 2-PHENYLETHYLAMINE, CALCIUM ION, GLYCEROL, ... | | Authors: | Leiros, H.-K.S, Brandsdal, B.O, Andersen, O.A, Os, V, Leiros, I, Helland, R, Otlewski, J, Willassen, N.P, Smalas, A.O. | | Deposit date: | 2003-12-09 | | Release date: | 2004-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Trypsin Specificity as Elucidated by Lie Calculations, X-Ray Structures, and Association Constant Measurements
Protein Sci., 13, 2004
|
|
2V28
 
 | | Apo structure of the cold active phenylalanine hydroxylase from Colwellia psychrerythraea 34H | | Descriptor: | PHENYLALANINE-4-HYDROXYLASE, SULFATE ION | | Authors: | Leiros, H.-K.S, Pey, A.L, Innselset, M, Moe, E, Leiros, I, Steen, I.H, Martinez, A. | | Deposit date: | 2007-06-04 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Phenylalanine Hydroxylase from Colwellia Psychrerythraea 34H, a Monomeric Cold Active Enzyme with Local Flexibility Around the Active Site and High Overall Stability.
J.Biol.Chem., 282, 2007
|
|
