7F28
 
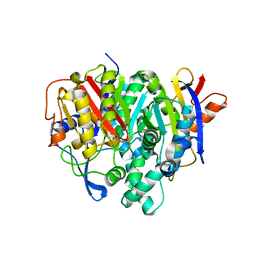 | | Crystal structure of a bacterial ketosynthase | | Descriptor: | Ketoacyl_synth_N domain-containing protein, Putative 3-oxoacyl-[ACP] synthase FabV | | Authors: | Lee, W.C, Kim, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | Structural basis of the complementary activity of two ketosynthases in aryl polyene biosynthesis.
Sci Rep, 11, 2021
|
|
2LGD
 
 | |
2MVF
 
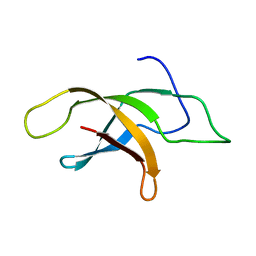 | | Structural insight into an essential assembly factor network on the pre-ribosome | | Descriptor: | Uncharacterized protein | | Authors: | Lee, W, Bassler, J, Paternoga, H, Holdermann, I, Thomas, M, Granneman, S, Barrio-Garcia, C, Nyarko, A, Stier, G, Clark, S.A, Schraivogel, D, Kallas, M, Beckmann, R, Tollervey, D, Barbar, E, Sinning, I, Hurt, E. | | Deposit date: | 2014-10-02 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation.
J.Cell Biol., 207, 2014
|
|
1G0V
 
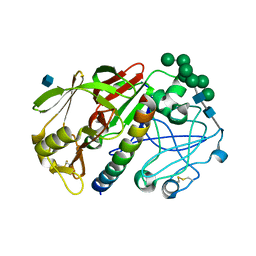 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH A IA3 MUTANT, MVV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEASE A INHIBITOR 3, PROTEINASE A, ... | | Authors: | Phylip, L.H, Lees, W, Brownsey, B.G, Bur, D, Dunn, B.M, Winther, J, Gustchina, A, Li, M, Copeland, T, Wlodawer, A, Kay, J. | | Deposit date: | 2000-10-09 | | Release date: | 2001-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The potency and specificity of the interaction between the IA3 inhibitor and its target aspartic proteinase from Saccharomyces cerevisiae.
J.Biol.Chem., 276, 2001
|
|
1FMU
 
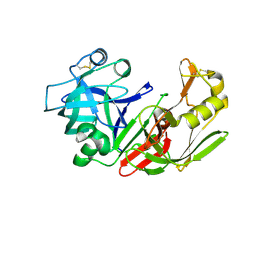 | | STRUCTURE OF NATIVE PROTEINASE A IN P3221 SPACE GROUP. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, SACCHAROPEPSIN, ... | | Authors: | Gustchina, A, Li, M, Phylip, L.H, Lees, W.E, Kay, J, Wlodawer, A. | | Deposit date: | 2000-08-18 | | Release date: | 2002-07-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An unusual orientation for Tyr75 in the active site of the aspartic proteinase from Saccharomyces cerevisiae.
Biochem.Biophys.Res.Commun., 295, 2002
|
|
1FMX
 
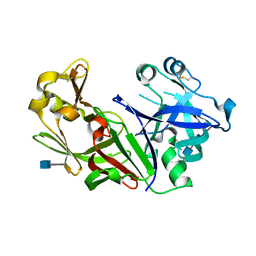 | | STRUCTURE OF NATIVE PROTEINASE A IN THE SPACE GROUP P21 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SACCHAROPEPSIN, ... | | Authors: | Gustchina, A, Li, M, Phylip, L.H, Lees, W.E, Kay, J, Wlodawer, A. | | Deposit date: | 2000-08-18 | | Release date: | 2002-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | An unusual orientation for Tyr75 in the active site of the aspartic proteinase from Saccharomyces cerevisiae.
Biochem.Biophys.Res.Commun., 295, 2002
|
|
1DPJ
 
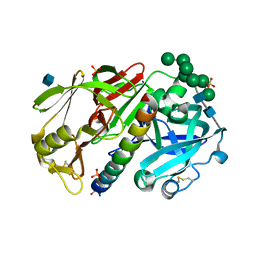 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH IA3 PEPTIDE INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEINASE A, PROTEINASE INHIBITOR IA3 PEPTIDE, ... | | Authors: | Li, M, Phylip, H.L, Lees, W.E, Winther, J.R, Dunn, B.M, Wlodawer, A, Kay, J, Guschina, A. | | Deposit date: | 1999-12-27 | | Release date: | 2000-05-03 | | Last modified: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The aspartic proteinase from Saccharomyces cerevisiae folds its own inhibitor into a helix.
Nat.Struct.Biol., 7, 2000
|
|
1DP5
 
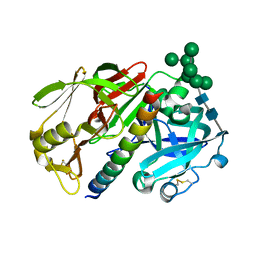 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH A IA3 MUTANT INHIBITOR | | Descriptor: | PROTEINASE A, PROTEINASE INHIBITOR IA3, beta-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, M, Phylip, H.L, Lees, W.E, Winther, J.R, Dunn, B.M, Wlodawer, A, Kay, J, Guschina, A. | | Deposit date: | 1999-12-23 | | Release date: | 2000-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The aspartic proteinase from Saccharomyces cerevisiae folds its own inhibitor into a helix.
Nat.Struct.Biol., 7, 2000
|
|
1MBB
 
 | | OXIDOREDUCTASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, URIDINE DIPHOSPHO-N-ACETYLENOLPYRUVYLGLUCOSAMINE REDUCTASE, URIDINE-DIPHOSPHATE-3(N-ACETYLGLUCOSAMINYL)BUTYRIC ACID | | Authors: | Benson, T.E, Lees, W.J, Walsh, C.T, Hogle, J.M. | | Deposit date: | 1995-11-07 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | (E)-enolbutyryl-UDP-N-acetylglucosamine as a mechanistic probe of UDP-N-acetylenolpyruvylglucosamine reductase (MurB).
Biochemistry, 35, 1996
|
|
5J4F
 
 | | Crystal structure of the N-terminally His6-tagged HP0902, an uncharacterized protein from Helicobacter pylori 26695 | | Descriptor: | Uncharacterized protein | | Authors: | Sim, D.-W, Lee, W.-C, Kim, H.Y, Kim, J.-H, Won, H.-S. | | Deposit date: | 2016-04-01 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural identification of the lipopolysaccharide-binding capability of a cupin-family protein from Helicobacter pylori
FEBS Lett., 590, 2016
|
|
2D38
 
 | | The Crystal Structure of Flavin Reductase HpaC complexed with NADP+ | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
2D36
 
 | | The Crystal Structure of Flavin Reductase HpaC | | Descriptor: | FLAVIN MONONUCLEOTIDE, hypothetical NADH-dependent FMN oxidoreductase | | Authors: | Okai, M, Kudo, N, Lee, W.C, Kamo, M, Nagata, K, Tanokura, M. | | Deposit date: | 2005-09-26 | | Release date: | 2006-05-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the short-chain flavin reductase HpaC from Sulfolobus tokodaii strain 7 in its three states: NAD(P)(+)(-)free, NAD(+)(-)bound, and NADP(+)(-)bound
Biochemistry, 45, 2006
|
|
3NGX
 
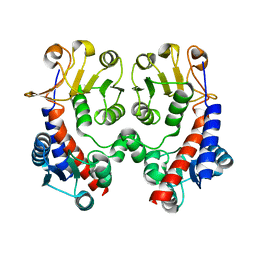 | |
3NGL
 
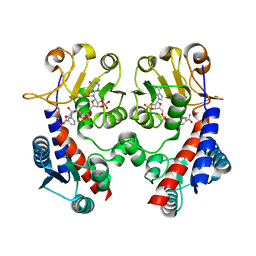 | | Crystal structure of bifunctional 5,10-methylenetetrahydrofolate dehydrogenase / cyclohydrolase from Thermoplasma acidophilum | | Descriptor: | Bifunctional protein folD, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Sung, M.W, Lee, W.H, Hwang, K.Y. | | Deposit date: | 2010-06-12 | | Release date: | 2011-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of bifunctional 5,10-methylenetetrahydrofolate dehydrogenase/cyclohydrolase from Thermoplasma acidophilum
Biochem.Biophys.Res.Commun., 406, 2011
|
|
5B6M
 
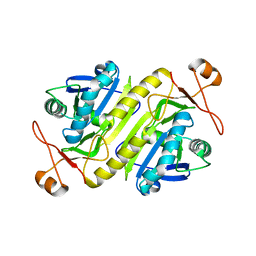 | |
6T8W
 
 | | Complement factor B in complex with (-)-4-(1-((5,7-Dimethyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic acid | | Descriptor: | 5,7-dimethyl-4-[[(2~{S})-2-phenylpiperidin-1-yl]methyl]-1~{H}-indole, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Sweeney, A.M, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Wu, M.S, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, Erkenez, A.D, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
6T8U
 
 | | Complement factor B in complex with 5-Bromo-3-chloro-N-(4,5-dihydro-1H-imidazol-2-yl)-7-methyl-1H-indol-4-amine | | Descriptor: | 5-bromanyl-3-chloranyl-~{N}-(1~{H}-imidazol-2-yl)-7-methyl-1~{H}-indol-4-amine, Complement factor B, SULFATE ION | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
6T8V
 
 | | Complement factor B in complex with (S)-5,7-Dimethyl-4-((2-phenylpiperidin-1-yl)methyl)-1H-indole | | Descriptor: | 4-[(2~{S})-1-[(5,7-dimethyl-1~{H}-indol-4-yl)methyl]piperidin-2-yl]benzoic acid, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
5J4G
 
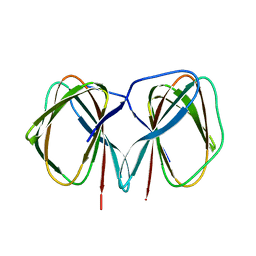 | | Crystal structure of the C-terminally His6-tagged HP0902, an uncharacterized protein from Helicobacter pylori 26695 | | Descriptor: | Uncharacterized protein | | Authors: | Sim, D.W, Lee, W.C, Kim, H.Y, Kim, J.H, Won, H.S. | | Deposit date: | 2016-04-01 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural identification of the lipopolysaccharide-binding capability of a cupin-family protein from Helicobacter pylori
FEBS Lett., 590, 2016
|
|
1MNL
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF A SWEET PROTEIN SINGLE-CHAIN MONELLIN (SCM) DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND DYNAMICAL SIMULATED ANNEALING CALCULATIONS, 21 STRUCTURES | | Descriptor: | MONELLIN | | Authors: | Lee, S.-Y, Lee, J.-H, Chang, H.-J, Jo, J.-M, Jung, J.-W, Lee, W. | | Deposit date: | 1998-08-06 | | Release date: | 1999-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a sweet protein single-chain monellin determined by nuclear magnetic resonance and dynamical simulated annealing calculations.
Biochemistry, 38, 1999
|
|
1EJQ
 
 | | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN IN THE PRESENCE OF PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE | | Descriptor: | SYNDECAN-4 | | Authors: | Shin, J, Oh, E.S, Lee, D, Couchman, J.R, Lee, W. | | Deposit date: | 2000-03-04 | | Release date: | 2001-03-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN IN THE PRESENCE OF PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE
To be Published
|
|
1EJP
 
 | | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN | | Descriptor: | SYNDECAN-4 | | Authors: | Lee, D, Oh, E.S, Woods, A, Couchman, J.R, Lee, W. | | Deposit date: | 2000-03-03 | | Release date: | 2001-09-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the dimeric cytoplasmic domain of syndecan-4.
Biochemistry, 40, 2001
|
|
6X6N
 
 | |
6O6W
 
 | | Solution structure of human myeloid-derived growth factor | | Descriptor: | Myeloid-derived growth factor | | Authors: | Bortnov, V, Tonelli, M, Lee, W, Markley, J.L, Mosher, D.F. | | Deposit date: | 2019-03-07 | | Release date: | 2019-11-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human myeloid-derived growth factor suggests a conserved function in the endoplasmic reticulum.
Nat Commun, 10, 2019
|
|
6WQE
 
 | | Solution Structure of the IWP-051-bound H-NOX from Shewanella woodyi in the Fe(II)CO ligation state | | Descriptor: | 5-fluoro-2-{1-[(2-fluorophenyl)methyl]-5-(1,2-oxazol-3-yl)-1H-pyrazol-3-yl}pyrimidin-4-ol, CARBON MONOXIDE, Heme NO binding domain protein, ... | | Authors: | Chen, C.Y, Lee, W, Montfort, W.R. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the Shewanella woodyi H-NOX protein in the presence and absence of soluble guanylyl cyclase stimulator IWP-051.
Protein Sci., 30, 2021
|
|
