9D6F
 
 | |
5TOD
 
 | | Transmembrane protein 24 SMP domain | | Descriptor: | Transmembrane protein 24 | | Authors: | Lees, J.A, Reinisch, K.M. | | Deposit date: | 2016-10-17 | | Release date: | 2017-03-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Lipid transport by TMEM24 at ER-plasma membrane contacts regulates pulsatile insulin secretion.
Science, 355, 2017
|
|
6BQ1
 
 | | Human PI4KIIIa lipid kinase complex | | Descriptor: | 5-{2-amino-1-[4-(morpholin-4-yl)phenyl]-1H-benzimidazol-6-yl}-N-(2-fluorophenyl)-2-methoxypyridine-3-sulfonamide, Phosphatidylinositol 4-kinase III alpha (PI4KA), Protein FAM126A, ... | | Authors: | Lees, J.A, Zhang, Y, Oh, M, Schauder, C.M, Yu, X, Baskin, J, Dobbs, K, Notarangelo, L.D, Camilli, P.D, Walz, T, Reinisch, K.M. | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of the human PI4KIII alpha lipid kinase complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7UJA
 
 | | Cryo-EM structure of Human respiratory syncytial virus F variant (construct pXCS847A) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AM14 Fab heavy chain, AM14 Fab light chain, ... | | Authors: | Lees, J.A, Ammirati, M, Han, S. | | Deposit date: | 2022-03-30 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Rational design of a highly immunogenic prefusion-stabilized F glycoprotein antigen for a respiratory syncytial virus vaccine.
Sci Transl Med, 15, 2023
|
|
8TB7
 
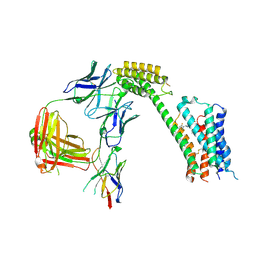 | | Cryo-EM Structure of GPR61- | | Descriptor: | 6-{[(3,5-difluoropyridin-4-yl)methyl]amino}-N-(4-ethoxy-6-methylpyrimidin-2-yl)-2-methoxy-N-(2-methoxyethyl)pyridine-3-sulfonamide, Fab hinge-binding nanobody, Fab24 BAK5 heavy chain, ... | | Authors: | Lees, J.A, Dias, J.M, Han, S. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | An inverse agonist of orphan receptor GPR61 acts by a G protein-competitive allosteric mechanism.
Nat Commun, 14, 2023
|
|
8TB0
 
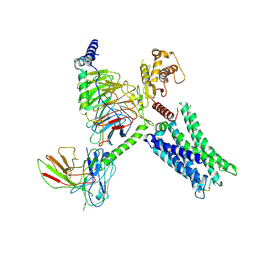 | | Cryo-EM Structure of GPR61-G protein complex stabilized by scFv16 | | Descriptor: | GPR61 fused to dominant negative G alpha S/I N18 chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Lees, J.A, Dias, J.M, Han, S. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | An inverse agonist of orphan receptor GPR61 acts by a G protein-competitive allosteric mechanism.
Nat Commun, 14, 2023
|
|
8V93
 
 | |
7L7F
 
 | |
7L7K
 
 | |
7K1W
 
 | |
7K2V
 
 | |
7K1Y
 
 | |
8WP9
 
 | | Small-heat shock protein from Methanocaldococcus jannaschii, Hsp16.5 | | Descriptor: | Small heat shock protein HSP16.5 | | Authors: | Lee, J, Ryu, B, Kim, T, Kim, K.K. | | Deposit date: | 2023-10-09 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM structure of a 16.5-kDa small heat-shock protein from Methanocaldococcus jannaschii.
Int.J.Biol.Macromol., 258, 2024
|
|
8GLZ
 
 | | Crystal structure of T252E-CYP199A4 in complex with 4-hydroxybenzoic acid. Crystal was initially co-crystallised with 4-methoxybenzoic acid and soaked with 4 mM hydrogen peroxide | | Descriptor: | CHLORIDE ION, Cytochrome P450, P-HYDROXYBENZOIC ACID, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2023-03-23 | | Release date: | 2023-12-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | An In Crystallo Reaction with an Engineered Cytochrome P450 Peroxygenase.
Chemistry, 30, 2024
|
|
8GM2
 
 | |
6IFH
 
 | | Unphosphorylated Spo0F from Paenisporosarcina sp. TG-14 | | Descriptor: | MAGNESIUM ION, Sporulation initiation phosphotransferase F | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of unphosphorylated Spo0F from Paenisporosarcina sp. TG-14, a psychrophilic bacterium isolated from an Antarctic glacier
Biodesign, 6(4), 2019
|
|
8GLY
 
 | | Crystal structure of T252E-CYP199A4 in complex with 4-hydroxybenzoic acid | | Descriptor: | CHLORIDE ION, Cytochrome P450, P-HYDROXYBENZOIC ACID, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2023-03-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | An In Crystallo Reaction with an Engineered Cytochrome P450 Peroxygenase.
Chemistry, 30, 2024
|
|
8GM1
 
 | | Crystal structure of T252E-CYP199A4 in complex with 4-methoxybenzoic acid soaked with 1 mM hydrogen peroxide | | Descriptor: | 4-METHOXYBENZOIC ACID, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2023-03-24 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An In Crystallo Reaction with an Engineered Cytochrome P450 Peroxygenase.
Chemistry, 30, 2024
|
|
3DZE
 
 | | Crystal structure of bovine coupling Factor B bound with cadmium | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP synthase subunit s, mitochondrial, ... | | Authors: | Lee, J.K, Stroud, R.M, Belogrudov, G.I. | | Deposit date: | 2008-07-29 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of bovine mitochondrial factor B at 0.96-A resolution.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5Z2E
 
 | |
6INT
 
 | | xylose isomerase from Paenibacillus sp. R4 | | Descriptor: | CALCIUM ION, Xylose isomerase | | Authors: | Lee, J.H, Lee, C.W, Park, S. | | Deposit date: | 2018-10-26 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Crystal Structure and Functional Characterization of a Xylose Isomerase (PbXI) from the Psychrophilic Soil Microorganism, Paenibacillus sp.
J. Microbiol. Biotechnol., 29, 2019
|
|
1JYS
 
 | | Crystal Structure of E. coli MTA/AdoHcy Nucleosidase | | Descriptor: | ADENINE, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2001-09-13 | | Release date: | 2002-10-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of E. coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase reveals similarity to the purine nucleoside phosphorylases.
Structure, 9, 2001
|
|
3UYV
 
 | | Crystal structure of a glycosylated ice-binding protein (LeIBP) from Arctic yeast | | Descriptor: | Antifreeze protein, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Lee, J.H, Park, A.K, Do, H, Park, K.S, Moh, S.H, Chi, Y.M, Kim, H.J. | | Deposit date: | 2011-12-06 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis for the antifreeze activity of an ice-binding protein from an Arctic yeast.
J.Biol.Chem., 2012
|
|
6IYM
 
 | |
3LNN
 
 | | Crystal structure of ZneB from Cupriavidus metallidurans | | Descriptor: | Membrane fusion protein (MFP) heavy metal cation efflux ZneB (CzcB-like), ZINC ION | | Authors: | Lee, J.K, De Angelis, F, Miercke, L.J, Stroud, R.M, Vandenbussche, G, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2010-02-02 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Metal-induced conformational changes in ZneB suggest an active role of membrane fusion proteins in efflux resistance systems.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
