4OH9
 
 | | Crystal Structure of the human MST2 SARAH homodimer | | Descriptor: | Serine/threonine-protein kinase 3 | | Authors: | Hwang, E, Cheong, H.-K, Ul Mushtaq, A, Kim, H.-Y, Yeo, K.J, Kim, E, Lee, W.C, Hwang, K.Y, Cheong, C, Jeon, Y.H. | | Deposit date: | 2014-01-17 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural basis of the heterodimerization of the MST and RASSF SARAH domains in the Hippo signalling pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OH8
 
 | | Crystal Structure of the human MST1-RASSF5 SARAH heterodimer | | Descriptor: | Ras association domain-containing protein 5, Serine/threonine-protein kinase 4 | | Authors: | Hwang, E, Cheong, H.-K, Ul Mushtaq, A, Kim, H.-Y, Yeo, K.J, Kim, E, Lee, W.C, Hwang, K.Y, Cheong, C, Jeon, Y.H. | | Deposit date: | 2014-01-17 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Structural basis of the heterodimerization of the MST and RASSF SARAH domains in the Hippo signalling pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3TRS
 
 | | The crystal structure of aspergilloglutamic peptidase from Aspergillus niger | | Descriptor: | Aspergillopepsin-2 heavy chain, Aspergillopepsin-2 light chain, DIMETHYL SULFOXIDE | | Authors: | Sasaki, H, Kubota, K, Lee, W.C, Ohtsuka, J, Kojima, M, Takahashi, K, Tanokura, M. | | Deposit date: | 2011-09-10 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of an intermediate dimer of aspergilloglutamic peptidase that mimics the enzyme-activation product complex produced upon autoproteolysis.
J.Biochem., 152, 2012
|
|
2MGQ
 
 | | Structure of CEH37 Homeodomain | | Descriptor: | Homeobox protein ceh-37 | | Authors: | Moon, S, Lee, W. | | Deposit date: | 2013-11-04 | | Release date: | 2014-11-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of CEH37 Homeodomain
To be Published
|
|
7EOY
 
 | | Engineered Hepatitis B virus core antigen T=3 | | Descriptor: | Capsid protein,Immunoglobulin G-binding protein A | | Authors: | Jeong, H, Heo, Y, Yoo, Y, Ryu, B, Yun, J, Cho, H, Lee, W. | | Deposit date: | 2021-04-24 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and Functional Characterizations of Cancer Targeting Nanoparticles Based on Hepatitis B Virus Capsid.
Int J Mol Sci, 22, 2021
|
|
7FDJ
 
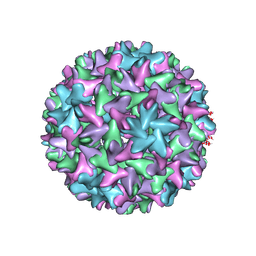 | | Engineered Hepatitis B virus core antigen with short linker T=4 | | Descriptor: | Capsid protein,Immunoglobulin G-binding protein A | | Authors: | Jeong, H, Heo, Y, Yoo, Y, Ryu, B, Yun, J, Cho, H, Lee, W. | | Deposit date: | 2021-07-16 | | Release date: | 2021-09-01 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural and Functional Characterizations of Cancer Targeting Nanoparticles Based on Hepatitis B Virus Capsid.
Int J Mol Sci, 22, 2021
|
|
7EP6
 
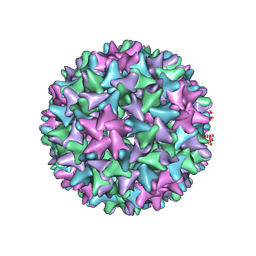 | | Engineered Hepatitis B virus core antigen T=4 | | Descriptor: | Capsid protein,Immunoglobulin G-binding protein A | | Authors: | Jeong, H, Heo, Y, Yoo, Y, Ryu, B, Yun, J, Cho, H, Lee, W. | | Deposit date: | 2021-04-26 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structural and Functional Characterizations of Cancer Targeting Nanoparticles Based on Hepatitis B Virus Capsid.
Int J Mol Sci, 22, 2021
|
|
6O6W
 
 | | Solution structure of human myeloid-derived growth factor | | Descriptor: | Myeloid-derived growth factor | | Authors: | Bortnov, V, Tonelli, M, Lee, W, Markley, J.L, Mosher, D.F. | | Deposit date: | 2019-03-07 | | Release date: | 2019-11-13 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human myeloid-derived growth factor suggests a conserved function in the endoplasmic reticulum.
Nat Commun, 10, 2019
|
|
7EB1
 
 | | Solution NMR structure of the RRM domain of RNA binding protein RBM3 from homo sapiens | | Descriptor: | RNA-binding protein 3 | | Authors: | Boral, S, Roy, S, Basak, A.J, Maiti, S, Lee, W, De, S. | | Deposit date: | 2021-03-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamic studies of the human RNA binding protein RBM3 reveals the molecular basis of its oligomerization and RNA recognition.
Febs J., 289, 2022
|
|
7W7Q
 
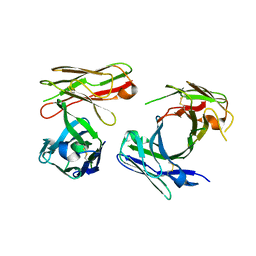 | |
6OCV
 
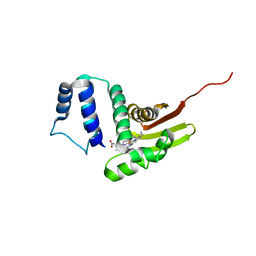 | |
1YY6
 
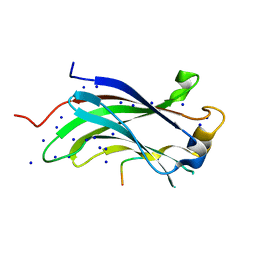 | | The Crystal Structure of the N-terminal domain of HAUSP/USP7 complexed with an EBNA1 peptide | | Descriptor: | Epstein-Barr nuclear antigen-1, SODIUM ION, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Saridakis, V, Sheng, Y, Sarkari, F, Holowaty, M, Shire, K, Nguyen, T, Zhang, R, Liao, J, Lee, W, Edwards, A.M, Arrowsmith, C.H, Frappier, L. | | Deposit date: | 2005-02-23 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization.
Mol.Cell, 18, 2005
|
|
1GMZ
 
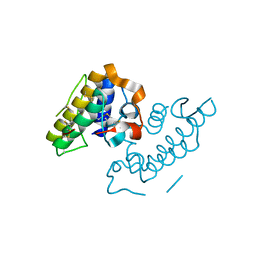 | |
7ELK
 
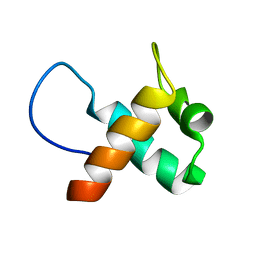 | |
7FCI
 
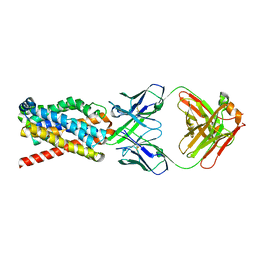 | | human NTCP in complex with YN69083 Fab | | Descriptor: | Fab Heavy chain, Fab Light chain, Sodium/bile acid cotransporter | | Authors: | Park, J.H, Iwamoto, M, Yun, J.H, Uchikubo-Kamo, T, Son, D, Jin, Z, Yoshida, H, Ohki, M, Ishimoto, N, Mizutani, K, Oshima, M, Muramatsu, M, Wakita, T, Shirouzu, M, Liu, K, Uemura, T, Nomura, N, Iwata, S, Watashi, K, Tame, J.R.H, Nishizawa, T, Lee, W, Park, S.Y. | | Deposit date: | 2021-07-14 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the HBV receptor and bile acid transporter NTCP.
Nature, 606, 2022
|
|
7F27
 
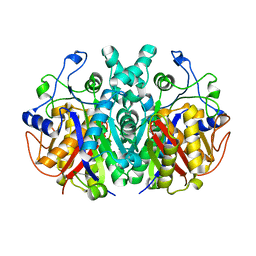 | | Crystal structure of polyketide ketosynthase | | Descriptor: | 3-oxoacyl-(Acyl-carrier-protein) synthase | | Authors: | Kim, Y, Lee, W.C. | | Deposit date: | 2021-06-10 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.806 Å) | | Cite: | Structural basis of the complementary activity of two ketosynthases in aryl polyene biosynthesis.
Sci Rep, 11, 2021
|
|
1YZE
 
 | | Crystal structure of the N-terminal domain of USP7/HAUSP. | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Saridakis, V, Sheng, Y, Sarkari, F, Holowaty, M.N, Shire, K, Nguyen, T, Zhang, R.G, Liao, J, Lee, W, Edwards, A.M, Arrowsmith, C.H, Frappier, L. | | Deposit date: | 2005-02-28 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization.
Mol.Cell, 18, 2005
|
|
2ABY
 
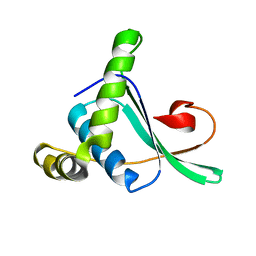 | | Solution structure of TA0743 from Thermoplasma acidophilum | | Descriptor: | hypothetical protein TA0743 | | Authors: | Kim, B, Jung, J, Hong, E, Yee, A, Arrowsmith, C.H, Lee, W. | | Deposit date: | 2005-07-18 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the conserved novel-fold protein TA0743 from Thermoplasma acidophilum.
Proteins, 62, 2006
|
|
6NF4
 
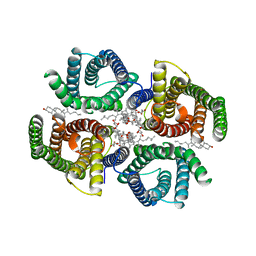 | | Structure of zebrafish Otop1 in nanodiscs | | Descriptor: | CHOLESTEROL, CHOLESTEROL HEMISUCCINATE, Otopetrin1 | | Authors: | Saotome, K, Lee, W.H, Liman, E.R, Ward, A.B. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structures of the otopetrin proton channels Otop1 and Otop3.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6NF6
 
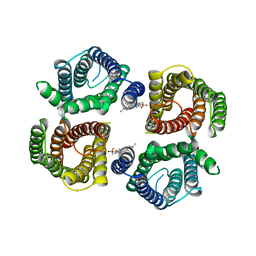 | | Structure of chicken Otop3 in nanodiscs | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Otopetrin3 | | Authors: | Saotome, K, Lee, W.H, Liman, E.R, Ward, A.B. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structures of the otopetrin proton channels Otop1 and Otop3.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2G3Y
 
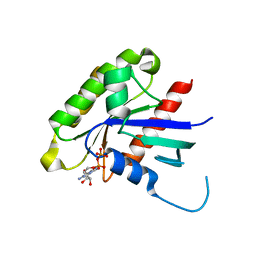 | | Crystal structure of the human small GTPase GEM | | Descriptor: | GTP-binding protein GEM, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Ugochukwu, E, Soundararajan, M, Elkins, J, Gileadi, C, Schoch, G, Sobott, F, Fedorov, O, Bray, J, Pantic, N, Berridge, G, Burgess, N, Lee, W.H, Turnbull, A, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, von Delft, F, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-02-21 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human small GTPase GEM
To be Published
|
|
1ZV4
 
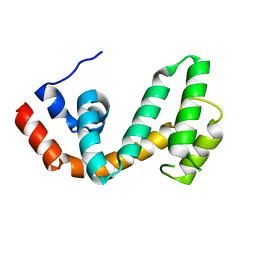 | | Structure of the Regulator of G-Protein Signaling 17 (RGSZ2) | | Descriptor: | Regulator of G-protein signaling 17 | | Authors: | Schoch, G.A, Jansson, A, Elkins, J.M, Haroniti, A, Niesen, F.H, Bunkoczi, G, Lee, W.H, Turnbull, A.P, Yang, X, Sundstrom, M, Arrowsmith, C, Edwards, A, Marsden, B, Gileadi, O, Ball, L, von Delft, F, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-01 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1FMU
 
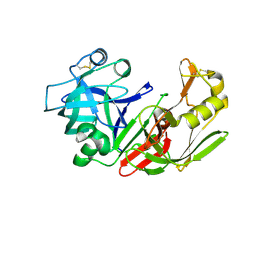 | | STRUCTURE OF NATIVE PROTEINASE A IN P3221 SPACE GROUP. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, SACCHAROPEPSIN, ... | | Authors: | Gustchina, A, Li, M, Phylip, L.H, Lees, W.E, Kay, J, Wlodawer, A. | | Deposit date: | 2000-08-18 | | Release date: | 2002-07-31 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An unusual orientation for Tyr75 in the active site of the aspartic proteinase from Saccharomyces cerevisiae.
Biochem.Biophys.Res.Commun., 295, 2002
|
|
1FMX
 
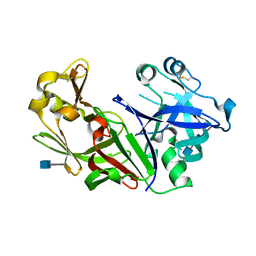 | | STRUCTURE OF NATIVE PROTEINASE A IN THE SPACE GROUP P21 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SACCHAROPEPSIN, ... | | Authors: | Gustchina, A, Li, M, Phylip, L.H, Lees, W.E, Kay, J, Wlodawer, A. | | Deposit date: | 2000-08-18 | | Release date: | 2002-07-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | An unusual orientation for Tyr75 in the active site of the aspartic proteinase from Saccharomyces cerevisiae.
Biochem.Biophys.Res.Commun., 295, 2002
|
|
1G0V
 
 | | THE STRUCTURE OF PROTEINASE A COMPLEXED WITH A IA3 MUTANT, MVV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEASE A INHIBITOR 3, PROTEINASE A, ... | | Authors: | Phylip, L.H, Lees, W, Brownsey, B.G, Bur, D, Dunn, B.M, Winther, J, Gustchina, A, Li, M, Copeland, T, Wlodawer, A, Kay, J. | | Deposit date: | 2000-10-09 | | Release date: | 2001-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The potency and specificity of the interaction between the IA3 inhibitor and its target aspartic proteinase from Saccharomyces cerevisiae.
J.Biol.Chem., 276, 2001
|
|
