2BH2
 
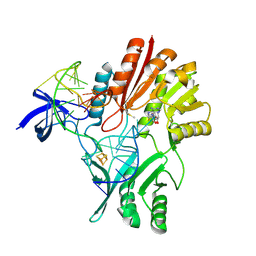 | | Crystal Structure of E. coli 5-methyluridine methyltransferase RumA in complex with ribosomal RNA substrate and S-adenosylhomocysteine. | | Descriptor: | 23S RIBOSOMAL RNA 1932-1968, 23S RRNA (URACIL-5-)-METHYLTRANSFERASE RUMA, IRON/SULFUR CLUSTER, ... | | Authors: | Lee, T.T, Agarwalla, S, Stroud, R.M. | | Deposit date: | 2005-01-06 | | Release date: | 2005-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Unique RNA Fold in the Ruma-RNA-Cofactor Ternary Complex Contributes to Substrate Selectivity and Enzymatic Function
Cell(Cambridge,Mass.), 120, 2005
|
|
2UYM
 
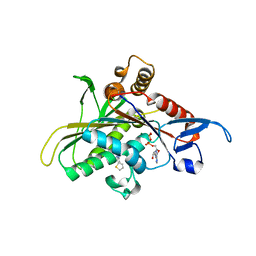 | |
2UYI
 
 | | Crystal structure of KSP in complex with ADP and thiophene containing inhibitor 33 | | Descriptor: | (5R)-N,N-DIETHYL-5-METHYL-2-[(THIOPHEN-2-YLCARBONYL)AMINO]-4,5,6,7-TETRAHYDRO-1-BENZOTHIOPHENE-3-CARBOXAMIDE, ADENOSINE-5'-DIPHOSPHATE, KINESIN-LIKE PROTEIN KIF11, ... | | Authors: | Lee, T.T. | | Deposit date: | 2007-04-07 | | Release date: | 2007-05-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis and Sar of Thiophene Containing Kinesin Spindle Protein (Ksp) Inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1UWV
 
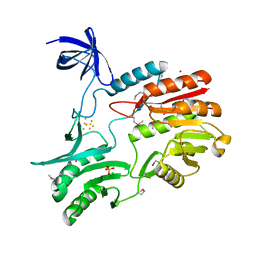 | | Crystal Structure of RumA, the iron-sulfur cluster containing E. coli 23S Ribosomal RNA 5-Methyluridine Methyltransferase | | Descriptor: | 23S RRNA (URACIL-5-)-METHYLTRANSFERASE RUMA, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Lee, T.T, Agarwalla, S, Stroud, R.M. | | Deposit date: | 2004-02-11 | | Release date: | 2004-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Ruma, an Iron-Sulfur Cluster Containing E. Coli Ribosomal RNA 5-Methyluridine Methyltransferase.
Structure, 12, 2004
|
|
2G8X
 
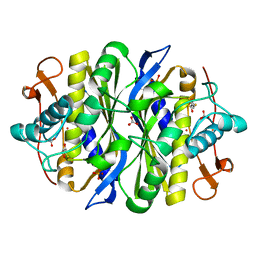 | | Escherichia coli Y209W apoprotein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CARBONATE ION, PHOSPHATE ION, ... | | Authors: | Lee, T.T, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2006-03-03 | | Release date: | 2006-03-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The role of protein dynamics in thymidylate synthase catalysis: Variants of conserved dUMP-binding Tyr-261
TO BE PUBLISHED
|
|
2PG2
 
 | |
6CDZ
 
 | | E. coli thymidylate synthase mutant I264Am | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2'-deoxy-5'-uridylic acid, ... | | Authors: | Finer-moore, J.S, Lee, T.T, Stroud, R.M. | | Deposit date: | 2018-02-09 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Single Mutation Traps a Half-Sites Reactive Enzyme in Midstream, Explaining Asymmetry in Hydride Transfer.
Biochemistry, 57, 2018
|
|
4GEV
 
 | | E. coli thymidylate synthase Y209W variant in complex with substrate and a cofactor analog | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-deoxy-5'-uridylic acid, Thymidylate synthase | | Authors: | Newby, Z, Lee, T.T, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2012-08-02 | | Release date: | 2012-08-29 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A remote mutation affects the hydride transfer by disrupting concerted protein motions in thymidylate synthase.
J.Am.Chem.Soc., 134, 2012
|
|
