4PGQ
 
 | |
4PHP
 
 | |
7FD5
 
 | | A complete three-dimensional structure of the Lon protease translocating a protein substrate (conformation 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Alpha-S1-casein, Lon protease, ... | | Authors: | Li, S, Hsieh, K, Kuo, C, Lee, S, Pintilie, G, Zhang, K, Chang, C. | | Deposit date: | 2021-07-16 | | Release date: | 2021-11-03 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Complete three-dimensional structures of the Lon protease translocating a protein substrate.
Sci Adv, 7, 2021
|
|
4M47
 
 | | structure of human DNA polymerase complexed with 8-BrG in the template base paired with incoming non-hydrolyzable GTP | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA polymerase beta, MAGNESIUM ION, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-06 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural basis for promutagenicity of 8-halogenated Guanine.
J.Biol.Chem., 289, 2014
|
|
4MF2
 
 | | Structure of human DNA polymerase beta complexed with O6MG as the template base in a 1-nucleotide gapped DNA | | Descriptor: | DNA polymerase beta, SODIUM ION, synthetic downstream primer, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-27 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Metal-dependent conformational activation explains highly promutagenic replication across O6-methylguanine by human DNA polymerase beta.
J.Am.Chem.Soc., 136, 2014
|
|
4NM2
 
 | | Structure of human DNA polymerase beta complexed with a nicked DNA containing a 8BrG-G at N-1 position and G-C at N position | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*GP*(BGM)P*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*GP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Koag, M.-C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.524 Å) | | Cite: | Structural basis for promutagenicity of 8-halogenated Guanine.
J.Biol.Chem., 289, 2014
|
|
4NLK
 
 | | Structure of human DNA polymerase beta complexed with 8BrG in the template base-paired with incoming non-hydrolyzable CTP | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, 5'-D(*CP*CP*GP*AP*CP*(BGM)P*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*A)-3', ... | | Authors: | Koag, M.-C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Structural basis for promutagenicity of 8-halogenated Guanine.
J.Biol.Chem., 289, 2014
|
|
4MF8
 
 | | Structure of human DNA polymerase beta complexed with nicked DNA containing a mismatched template O6MG and incoming CTP | | Descriptor: | DNA polymerase beta, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-27 | | Release date: | 2014-08-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structures of human DNA polymerase beta inserting bases opposite templating O6MG
To be Published
|
|
4MFF
 
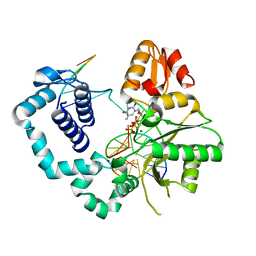 | | Structure of human DNA polymerase beta complexed with O6MG in the template base paired with incoming non-hydrolyzable TTP | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA polymerase beta, MAGNESIUM ION, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-27 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Metal-dependent conformational activation explains highly promutagenic replication across O6-methylguanine by human DNA polymerase beta.
J.Am.Chem.Soc., 136, 2014
|
|
4MFC
 
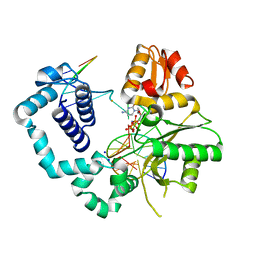 | | Structure of human DNA polymerase beta complexed with O6MG in the template base paired with incoming non-hydrolyzable CTP | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA polymerase beta, MAGNESIUM ION, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-27 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Metal-dependent conformational activation explains highly promutagenic replication across O6-methylguanine by human DNA polymerase beta.
J.Am.Chem.Soc., 136, 2014
|
|
4M2Y
 
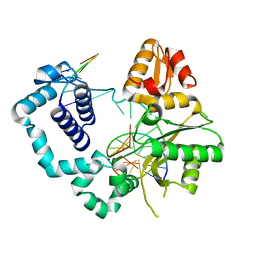 | | Structure of human DNA polymerase beta complexed with 8-BrG as the template base in a 1-nucleotide gapped DNA | | Descriptor: | DNA polymerase beta, SODIUM ION, down-primer, ... | | Authors: | Koag, M.C, Min, K, Monzingo, A.F, Lee, S. | | Deposit date: | 2013-08-05 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural basis for promutagenicity of 8-halogenated Guanine.
J.Biol.Chem., 289, 2014
|
|
5HHH
 
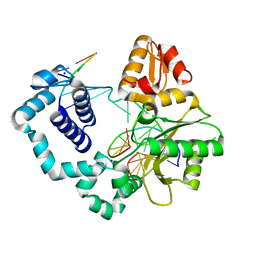 | | Structure of human DNA polymerase beta Host-Guest complexed with the control G for N7-CBZ-platination | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*AP*GP*GP*AP*GP*CP*AP*GP*G)-3'), DNA (5'-D(P*CP*CP*TP*GP*CP*TP*CP*CP*TP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Koag, M.-C, Lee, S. | | Deposit date: | 2016-01-11 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.363 Å) | | Cite: | Synthesis, structure, and biological evaluation of a platinum-carbazole conjugate.
Chem Biol Drug Des, 91, 2018
|
|
2F66
 
 | | Structure of the ESCRT-I endosomal trafficking complex | | Descriptor: | Protein SRN2, SULFATE ION, Suppressor protein STP22 of temperature-sensitive alpha-factor receptor and arginine permease, ... | | Authors: | Kostelansky, M.S, Lee, S, Kim, J, Hurley, J.H. | | Deposit date: | 2005-11-28 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional organization of the ESCRT-I trafficking complex.
Cell(Cambridge,Mass.), 125, 2006
|
|
7FDB
 
 | | CryoEM Structures of Reconstituted V-ATPase,State2 | | Descriptor: | Fusion of yeast V-type proton ATPase subunit H(NT) and human V-type proton ATPase subunit H(CT), V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
7FDA
 
 | | CryoEM Structure of Reconstituted V-ATPase, state1 | | Descriptor: | Fusion of yeast V-type proton ATPase subunit H(NT) and human V-type proton ATPase subunit H(CT), V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
7FDC
 
 | | CryoEM Structures of Reconstituted V-ATPase, state3 | | Descriptor: | Fusion of yeast V-type proton ATPase subunit H(NT) and human V-type proton ATPase subunit H(CT), V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
7FDE
 
 | | CryoEM Structures of Reconstituted V-ATPase, Oxr1 bound V1 | | Descriptor: | Oxidation resistance protein 1, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | Authors: | Khan, M.M, Lee, S, Oot, R.A, Couoh-Cardel, S, KIm, H, Wilkens, S, Roh, S.H. | | Deposit date: | 2021-07-16 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | CryoEM Structures of Reconstituted V-ATPase and Oxr1-bound V1 Reveal a Novel Mechanism of Regulation.
Embo J., 2021
|
|
2K3M
 
 | | Rv1761c | | Descriptor: | Rv1761c, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Page, R.C, Moore, J.D, Lee, S, Opella, S.J, Cross, T.A. | | Deposit date: | 2008-05-14 | | Release date: | 2009-01-06 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Backbone structure of a small helical integral membrane protein: A unique structural characterization.
Protein Sci., 18, 2009
|
|
2HNH
 
 | | Crystal structure of the catalytic alpha subunit of E. coli replicative DNA polymerase III | | Descriptor: | DNA polymerase III alpha subunit, PHOSPHATE ION | | Authors: | Meindert, M.H, Georgescu, R.E, Lee, S, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2006-07-12 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Catalytic alpha Subunit of E. coli Replicative DNA Polymerase III.
Cell(Cambridge,Mass.), 126, 2006
|
|
6JCM
 
 | | Crystal structure of ligand-free Rv0187. | | Descriptor: | ACETATE ION, Probable O-methyltransferase | | Authors: | Kim, J, Lee, S. | | Deposit date: | 2019-01-29 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural and biochemical characterization of Rv0187, an O-methyltransferase from Mycobacterium tuberculosis.
Sci Rep, 9, 2019
|
|
6JCL
 
 | | Crystal structure of cofactor-bound Rv0187 from MTB | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Probable O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Kim, J, Lee, S. | | Deposit date: | 2019-01-29 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.644 Å) | | Cite: | Structural and biochemical characterization of Rv0187, an O-methyltransferase from Mycobacterium tuberculosis.
Sci Rep, 9, 2019
|
|
2N8V
 
 | | An NMR/SAXS structure of the PKI domain of the honeybee dicistrovirus, Israeli acute paralysis virus (IAPV) IRES | | Descriptor: | RNA (70-MER) | | Authors: | Au, H.H, Cornilescu, G, Mouzakis, K.D, Burke, J.E, Ren, Q, Lee, S, Butcher, S.E, Jan, E. | | Deposit date: | 2015-10-27 | | Release date: | 2015-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Global shape mimicry of tRNA within a viral internal ribosome entry site mediates translational reading frame selection.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6LHS
 
 | | High resolution structure of FANCA C-terminal domain (CTD) | | Descriptor: | Fanconi anemia complementation group A | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
6LHU
 
 | | High resolution structure of FANCA C-terminal domain (CTD) | | Descriptor: | Fanconi anemia complementation group A | | Authors: | Jeong, E, Lee, S, Shin, J, Kim, Y, Kim, J, Scharer, O, Kim, Y, Kim, H, Cho, Y. | | Deposit date: | 2019-12-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis of the fanconi anemia-associated mutations within the FANCA and FANCG complex.
Nucleic Acids Res., 48, 2020
|
|
7KLF
 
 | |
