5VAQ
 
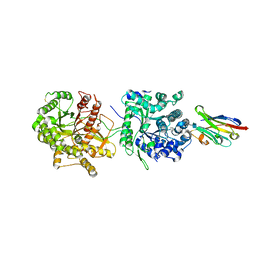 | | Crystal Structure of Beta-Klotho in Complex with FGF21CT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-klotho, Fibroblast growth factor 21, ... | | Authors: | Lee, S, Schlessinger, J. | | Deposit date: | 2017-03-27 | | Release date: | 2018-01-31 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Structures of beta-klotho reveal a 'zip code'-like mechanism for endocrine FGF signalling.
Nature, 553, 2018
|
|
3CRA
 
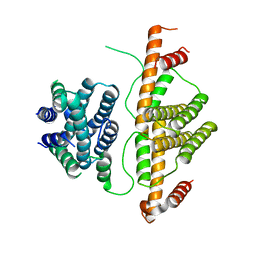 | | Crystal Structure of Escherichia coli MazG, the Regulator of Nutritional Stress Response | | Descriptor: | Protein mazG | | Authors: | Lee, S, Kim, M.H, Kang, B.S, Kim, J.S, Kim, Y.G, Kim, K.J. | | Deposit date: | 2008-04-05 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Escherichia coli MazG, the regulator of nutritional stress response.
J.Biol.Chem., 283, 2008
|
|
3CRC
 
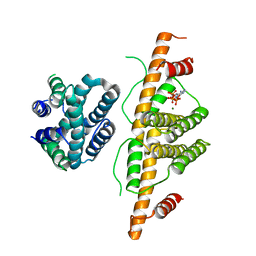 | | Crystal Structure of Escherichia coli MazG, the Regulator of Nutritional Stress Response | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein mazG | | Authors: | Lee, S, Kim, M.H, Kang, B.S, Kim, J.S, Kim, Y.G, Kim, K.J. | | Deposit date: | 2008-04-05 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Escherichia coli MazG, the regulator of nutritional stress response.
J.Biol.Chem., 283, 2008
|
|
3EVK
 
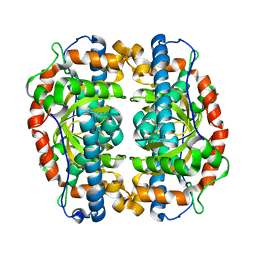 | |
3DAK
 
 | |
4P2H
 
 | |
5CUS
 
 | | Crystal Structure of sErbB3-Fab3379 Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab LC region of KTN3379, IgG H chain, ... | | Authors: | Lee, S, Schlessinger, J. | | Deposit date: | 2015-07-25 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Inhibition of ErbB3 by a monoclonal antibody that locks the extracellular domain in an inactive configuration.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6AVS
 
 | | Complex structure of JMJD5 and Symmetric Monomethyl-Arginine (MMA) | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, Lysine-specific demethylase 8, ZINC ION | | Authors: | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | Deposit date: | 2017-09-04 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
3PMR
 
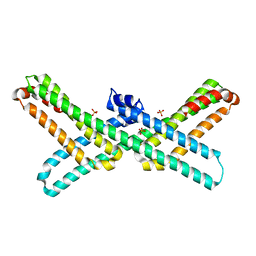 | | Crystal Structure of E2 domain of Human Amyloid Precursor-Like Protein 1 | | Descriptor: | Amyloid-like protein 1, PHOSPHATE ION | | Authors: | Lee, S, Xue, Y, Hu, J, Wang, Y, Liu, X, Demeler, B, Ha, Y. | | Deposit date: | 2010-11-17 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The E2 Domains of APP and APLP1 Share a Conserved Mode of Dimerization.
Biochemistry, 50, 2011
|
|
5HHI
 
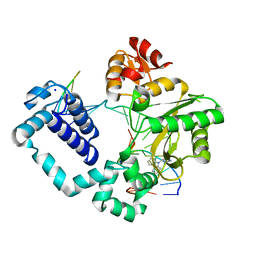 | | Structure of human DNA polymerase beta Host-Guest complexed with CBZ-platinated N7-G | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*AP*GP*GP*AP*GP*CP*AP*GP*G)-3'), DNA (5'-D(P*CP*CP*TP*GP*CP*TP*CP*CP*TP*C)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Lee, S, Koag, M.-C. | | Deposit date: | 2016-01-11 | | Release date: | 2017-01-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.517 Å) | | Cite: | Synthesis, structure, and biological evaluation of a platinum-carbazole conjugate.
Chem Biol Drug Des, 91, 2018
|
|
5IP5
 
 | |
4HYX
 
 | | Crystal Structure Analysis of the Bacteriorhodopsin in Facial Amphiphile-4 DMPC Bicelle | | Descriptor: | Bacteriorhodopsin, DECANE, GLYCEROL, ... | | Authors: | Lee, S, Stout, C.D, Zhang, Q. | | Deposit date: | 2012-11-14 | | Release date: | 2013-03-20 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HWL
 
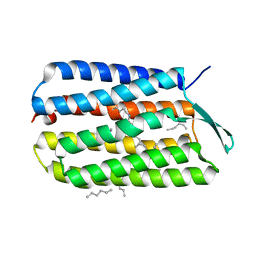 | | Crystal Structure Analysis of the Bacteriorhodopsin in Facial Amphiphile-7 DMPC Bicelle | | Descriptor: | Bacteriorhodopsin, GLYCEROL, HEPTANE, ... | | Authors: | Lee, S, Stout, C.D, Zhang, Q. | | Deposit date: | 2012-11-08 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2KMK
 
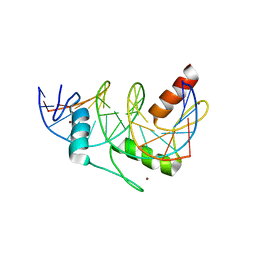 | | Gfi-1 Zinc Fingers 3-5 complexed with DNA | | Descriptor: | DNA (5'-D(*CP*AP*TP*AP*AP*AP*TP*CP*AP*CP*TP*GP*CP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*GP*CP*AP*GP*TP*GP*AP*TP*TP*TP*AP*TP*G)-3'), ZINC ION, ... | | Authors: | Lee, S, Wu, Z. | | Deposit date: | 2009-07-30 | | Release date: | 2010-03-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Gfi-1 zinc domain bound to consensus DNA.
J.Mol.Biol., 397, 2010
|
|
2FIF
 
 | |
2FID
 
 | |
5XD9
 
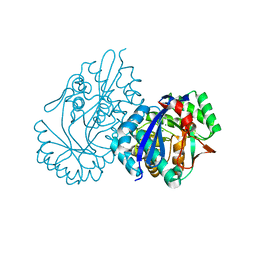 | | Crystal structure analysis of 3,6-anhydro-L-galactonate cycloisomerase | | Descriptor: | 3,6-anhydro-alpha-L-galactonate cycloisomerase, MAGNESIUM ION | | Authors: | Lee, S, Choi, I.-G, Kim, H.-Y. | | Deposit date: | 2017-03-27 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure analysis of 3,6-anhydro-l-galactonate cycloisomerase suggests emergence of novel substrate specificity in the enolase superfamily
Biochem. Biophys. Res. Commun., 491, 2017
|
|
5XD8
 
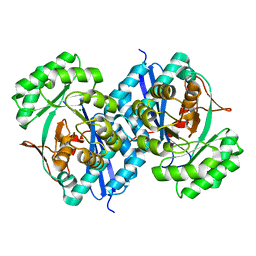 | | Crystal structure analysis of 3,6-anhydro-L-galactonate cycloisomerase | | Descriptor: | 3,6-anhydro-alpha-L-galactonate cycloisomerase, MAGNESIUM ION | | Authors: | Lee, S, Choi, I.-G, Kim, H.-Y. | | Deposit date: | 2017-03-27 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Crystal structure analysis of 3,6-anhydro-l-galactonate cycloisomerase suggests emergence of novel substrate specificity in the enolase superfamily
Biochem. Biophys. Res. Commun., 491, 2017
|
|
5XD7
 
 | | Crystal structure analysis of 3,6-anhydro-L-galactonate cycloisomerase | | Descriptor: | 3,6-anhydro-alpha-L-galactonate cycloisomerase, ACETIC ACID, MAGNESIUM ION | | Authors: | Lee, S, Choi, I.-G, Kim, H.-Y. | | Deposit date: | 2017-03-27 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Crystal structure analysis of 3,6-anhydro-l-galactonate cycloisomerase suggests emergence of novel substrate specificity in the enolase superfamily.
Biochem. Biophys. Res. Commun., 491, 2017
|
|
2OJQ
 
 | | Crystal structure of Alix V domain | | Descriptor: | Programmed cell death 6-interacting protein | | Authors: | Lee, S, Hurley, J.H. | | Deposit date: | 2007-01-13 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structural basis for viral late-domain binding to Alix
Nat.Struct.Mol.Biol., 14, 2007
|
|
4CSS
 
 | | Crystal structure of FimH in complex with a sulfonamide biphenyl alpha D-mannoside | | Descriptor: | 4'-(alpha-D-Mannopyranosyloxy)-biphenyl-4-methyl sulfonamide, GLYCEROL, PROTEIN FIMH | | Authors: | Kleeb, S, Pang, L, Mayer, K, Sigl, A, Eris, D, Preston, R.C, Zihlmann, P, Abgottspon, D, Hutter, A, Scharenberg, M, Jian, X, Navarra, G, Rabbani, S, Smiesko, M, Luedin, N, Jakob, R.P, Schwardt, O, Maier, T, Sharpe, T, Ernst, B. | | Deposit date: | 2014-03-10 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.069 Å) | | Cite: | Fimh Antagonists: Bioisosteres to Improve the in Vitro and in Vivo Pk/Pd Profile.
J.Med.Chem., 58, 2015
|
|
4CST
 
 | | Crystal structure of FimH in complex with 3'-Chloro-4'-(alpha-D-mannopyranosyloxy)-biphenyl-4-carbonitrile | | Descriptor: | 3'-chloro-4'-(alpha-D-mannopyranosyloxy)biphenyl-4-carbonitrile, PROTEIN FIMH | | Authors: | Kleeb, S, Pang, L, Mayer, K, Sigl, A, Eris, D, Preston, R.C, Zihlmann, P, Abgottspon, D, Hutter, A, Scharenberg, M, Jian, X, Navarra, G, Rabbani, S, Smiesko, M, Luedin, N, Jakob, R.P, Schwardt, O, Maier, T, Sharpe, T, Ernst, B. | | Deposit date: | 2014-03-10 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fimh Antagonists: Bioisosteres to Improve the in Vitro and in Vivo Pk/Pd Profile.
J.Med.Chem., 58, 2015
|
|
8J0J
 
 | | AtSLAC1 8D mutant in closed state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2023-04-11 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
8J1E
 
 | | AtSLAC1 in open state | | Descriptor: | CHLORIDE ION, CHOLESTEROL HEMISUCCINATE, Guard cell S-type anion channel SLAC1,Green fluorescent protein | | Authors: | Lee, Y, Lee, S. | | Deposit date: | 2023-04-12 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structures of the plant anion channel SLAC1 from Arabidopsis thaliana suggest a combined activation model.
Nat Commun, 14, 2023
|
|
3KL9
 
 | | Crystal structure of PepA from Streptococcus pneumoniae | | Descriptor: | Glutamyl aminopeptidase, ZINC ION | | Authors: | Kim, K.K, Lee, S, Kim, D. | | Deposit date: | 2009-11-07 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the substrate specificity of PepA from Streptococcus pneumoniae, a dodecameric tetrahedral protease
Biochem.Biophys.Res.Commun., 391, 2010
|
|
