5KUK
 
 | | Crystal Structure of Inward Rectifier Kir2.2 K62W Mutant | | Descriptor: | ATP-sensitive inward rectifier potassium channel 12, DECYL-BETA-D-MALTOPYRANOSIDE, POTASSIUM ION | | Authors: | Lee, S.-J, Ren, F, Heyman, S, Yuan, P, Nichols, C.G. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of control of inward rectifier Kir2 channel gating by bulk anionic phospholipids.
J.Gen.Physiol., 148, 2016
|
|
5ZCG
 
 | | Crystal structure of OsPP2C50 S265L/I267V:OsPYL/RCAR3 with (+)-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABA receptor RCAR3, MAGNESIUM ION, ... | | Authors: | Lee, S, Han, S. | | Deposit date: | 2018-02-17 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Comprehensive survey of the VxG Phi L motif of PP2Cs from Oryza sativa reveals the critical role of the fourth position in regulation of ABA responsiveness.
Plant Mol.Biol., 101, 2019
|
|
5KUM
 
 | | Crystal Structure of Inward Rectifier Kir2.2 K62W Mutant In Complex with PIP2 | | Descriptor: | ATP-sensitive inward rectifier potassium channel 12, DECYL-BETA-D-MALTOPYRANOSIDE, POTASSIUM ION, ... | | Authors: | Lee, S.-J, Ren, F, Heyman, S, Yuan, P, Nichols, C.G. | | Deposit date: | 2016-07-13 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of control of inward rectifier Kir2 channel gating by bulk anionic phospholipids.
J.Gen.Physiol., 148, 2016
|
|
3NIO
 
 | | Crystal structure of Pseudomonas aeruginosa guanidinobutyrase | | Descriptor: | Guanidinobutyrase, MANGANESE (II) ION | | Authors: | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Jang, J.Y, Im, H, An, D, Suh, S.W. | | Deposit date: | 2010-06-16 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of Pseudomonas aeruginosa guanidinobutyrase and guanidinopropionase, members of the ureohydrolase superfamily
J.Struct.Biol., 175, 2011
|
|
5ZCL
 
 | | Crystal structure of OsPP2C50 I267L:OsPYL/RCAR3 with (+)-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABA receptor RCAR3, MAGNESIUM ION, ... | | Authors: | Lee, S, Han, S. | | Deposit date: | 2018-02-19 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.661 Å) | | Cite: | Comprehensive survey of the VxG Phi L motif of PP2Cs from Oryza sativa reveals the critical role of the fourth position in regulation of ABA responsiveness.
Plant Mol.Biol., 101, 2019
|
|
3NIP
 
 | | Crystal structure of Pseudomonas aeruginosa guanidinopropionase complexed with 1,6-diaminohexane | | Descriptor: | 3-guanidinopropionase, HEXANE-1,6-DIAMINE | | Authors: | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Jang, J.Y, Im, H, An, D, Suh, S.W. | | Deposit date: | 2010-06-16 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Pseudomonas aeruginosa guanidinobutyrase and guanidinopropionase, members of the ureohydrolase superfamily
J.Struct.Biol., 175, 2011
|
|
4Q25
 
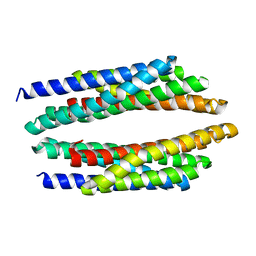 | | Crystal structure of PhoU from Pseudomonas aeruginosa | | Descriptor: | Phosphate-specific transport system accessory protein PhoU homolog | | Authors: | Lee, S.J, Lee, B.-J, Suh, S.W. | | Deposit date: | 2014-04-07 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of PhoU from Pseudomonas aeruginosa, a negative regulator of the Pho regulon.
J.Struct.Biol., 188, 2014
|
|
3NIQ
 
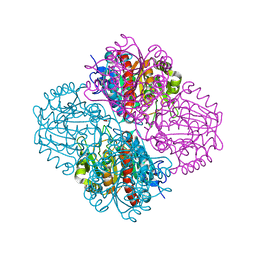 | | Crystal structure of Pseudomonas aeruginosa guanidinopropionase | | Descriptor: | 3-guanidinopropionase, GLYCEROL, MANGANESE (II) ION | | Authors: | Lee, S.J, Kim, H.S, Kim, D.J, Yoon, H.J, Kim, K.H, Yoon, J.Y, Jang, J.Y, Im, H, An, D, Suh, S.W. | | Deposit date: | 2010-06-16 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structures of Pseudomonas aeruginosa guanidinobutyrase and guanidinopropionase, members of the ureohydrolase superfamily
J.Struct.Biol., 175, 2011
|
|
5GHV
 
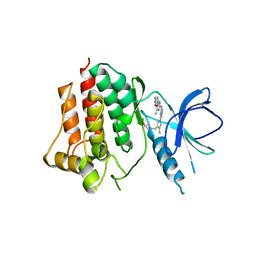 | | Crystal structure of an inhibitor-bound Syk | | Descriptor: | 1-({1-[2-({3,5-dimethyl-4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}amino)pyrimidin-4-yl]-4-methyl-1H-pyrrol-3-yl}methyl)azetidin-3-ol, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
5Y5U
 
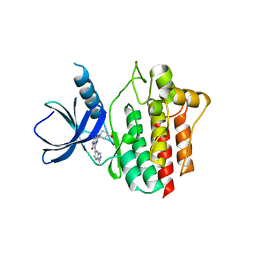 | | Crystal structures of spleen tyrosine kinase in complex with a novel inhibitor | | Descriptor: | 4-[(1-methylindazol-5-yl)amino]-2-(4-oxidanylpiperidin-1-yl)-8H-pyrido[4,3-d]pyrimidin-5-one, Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Lee, B.I. | | Deposit date: | 2017-08-09 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal Structures of Spleen Tyrosine Kinase in Complex with Two Novel 4-Aminopyrido[4,3-d] Pyrimidine Derivative Inhibitors.
Mol. Cells, 41, 2018
|
|
2KM9
 
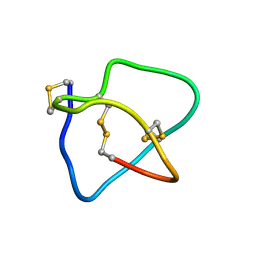 | | Omega conotoxin-FVIA | | Descriptor: | omega_conotoxin-FVIA | | Authors: | Lee, S, Kim, J, Lee, J, Jung, H. | | Deposit date: | 2009-07-25 | | Release date: | 2010-07-28 | | Last modified: | 2011-09-28 | | Method: | SOLUTION NMR | | Cite: | Structure of omega conotoxin-FVIA
To be Published
|
|
6M86
 
 | |
8QH5
 
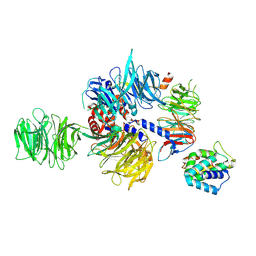 | | CryoEM structure of UVSSA(VHS)-CSA-DDB1-DDA1 | | Descriptor: | DET1- and DDB1-associated protein 1, DNA damage-binding protein 1, DNA excision repair protein ERCC-8, ... | | Authors: | Lee, S.-H, Sixma, T.K. | | Deposit date: | 2023-09-06 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CryoEM structure of UVSSA(VHS)-CSA-DDB1-DDA1
To Be Published
|
|
6LRP
 
 | |
8H5M
 
 | |
8H5K
 
 | |
8H5O
 
 | |
8H5J
 
 | |
8H5L
 
 | |
7W1G
 
 | | Crystal structure of YfiH with C107A mutation in complex with UDP-MurNAc-L-Serine | | Descriptor: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, (2S)-2-[[(2R)-2-[(2R,3R,4R,5S,6R)-3-acetamido-2-[[[(2R,3S,4R,5R)-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-6-(hydroxymethyl)-5-oxidanyl-oxan-4-yl]oxypropanoyl]amino]-3-oxidanyl-propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Lee, S.H, Hsieh, K.Y, Lee, M.S, Chang, C.I. | | Deposit date: | 2021-11-19 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Basis for the Peptidoglycan-Editing Activity of YfiH.
Mbio, 13, 2021
|
|
6O86
 
 | |
6O88
 
 | | Crystal Structure of UDP-dependent glucosyltransferases (UGT) from Stevia rebaudiana in complex with UDP and rebaudioside A | | Descriptor: | (8alpha,9beta,10alpha,13alpha)-13-{[alpha-L-allopyranosyl-(1->2)-[beta-D-mannopyranosyl-(1->3)]-beta-D-allopyranosyl]oxy}kauran-18-oic acid, UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Lee, S.G, Jez, J.M. | | Deposit date: | 2019-03-09 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular basis for branched steviol glucoside biosynthesis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3QHP
 
 | |
5ZCH
 
 | | Crystal structure of OsPP2C50 I267W:OsPYL/RCAR3 with (+)-ABA | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYL3, MAGNESIUM ION, ... | | Authors: | Lee, S, Han, S. | | Deposit date: | 2018-02-17 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.474 Å) | | Cite: | Comprehensive survey of the VxG Phi L motif of PP2Cs from Oryza sativa reveals the critical role of the fourth position in regulation of ABA responsiveness.
Plant Mol.Biol., 101, 2019
|
|
3R4Y
 
 | | Crystal structure of alpha-neoagarobiose hydrolase (ALPHA-NABH) from Saccharophagus degradans 2-40 | | Descriptor: | Glycosyl hydrolase family 32, N terminal | | Authors: | Lee, S, Lee, J.Y, Ha, S.C, Shin, D.H, Kim, K.H, Bang, W.G, Kim, S.H, Choi, I.G. | | Deposit date: | 2011-03-18 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a key enzyme in the agarolytic pathway, alpha-neoagarobiose hydrolase from Saccharophagus degradans 2-40
Biochem.Biophys.Res.Commun., 412, 2011
|
|
