5WPM
 
 | | KRas G12V, bound to GppNHp and miniprotein 225-11(A30R) | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Lee, S.-J, Shim, S.Y, McGee, J.H, Verdine, G.L. | | Deposit date: | 2017-08-05 | | Release date: | 2018-01-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Exceptionally high-affinity Ras binders that remodel its effector domain.
J. Biol. Chem., 293, 2018
|
|
5WPL
 
 | | KRas G12V, bound to GppNHp and miniprotein 225-11 | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Lee, S.-J, Shim, S.Y, McGee, J.H, Verdine, G.L. | | Deposit date: | 2017-08-05 | | Release date: | 2018-01-03 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Exceptionally high-affinity Ras binders that remodel its effector domain.
J. Biol. Chem., 293, 2018
|
|
1MNL
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF A SWEET PROTEIN SINGLE-CHAIN MONELLIN (SCM) DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND DYNAMICAL SIMULATED ANNEALING CALCULATIONS, 21 STRUCTURES | | Descriptor: | MONELLIN | | Authors: | Lee, S.-Y, Lee, J.-H, Chang, H.-J, Jo, J.-M, Jung, J.-W, Lee, W. | | Deposit date: | 1998-08-06 | | Release date: | 1999-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a sweet protein single-chain monellin determined by nuclear magnetic resonance and dynamical simulated annealing calculations.
Biochemistry, 38, 1999
|
|
6M2D
 
 | | MUL1-RING domain | | Descriptor: | Mitochondrial ubiquitin ligase activator of NFKB 1, SULFATE ION, ZINC ION | | Authors: | Lee, S.O, Ryu, K.S, Chi, S.-W. | | Deposit date: | 2020-02-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | MUL1-RING recruits the substrate, p53-TAD as a complex with UBE2D2-UB conjugate.
Febs J., 2022
|
|
6M2C
 
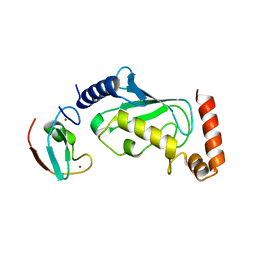 | |
5WP5
 
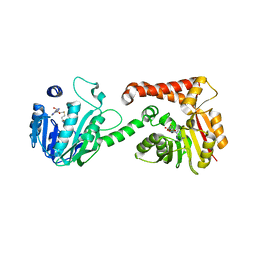 | |
5WP4
 
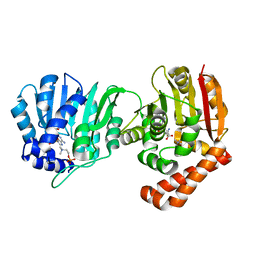 | |
6NZ5
 
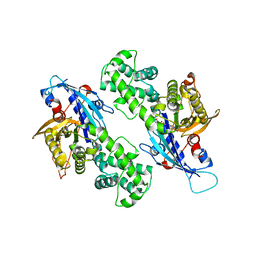 | | YcjX-GDPCP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, YcjX Stress Protein | | Authors: | Lee, S, Tsai, J, Tsai, F.T, Sung, N, Lee, J, Chang, C. | | Deposit date: | 2019-02-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | Crystal Structure of the YcjX Stress Protein Reveals a Ras-Like GTP-Binding Protein.
J.Mol.Biol., 431, 2019
|
|
1SVP
 
 | | SINDBIS VIRUS CAPSID PROTEIN | | Descriptor: | SINDBIS VIRUS CAPSID PROTEIN | | Authors: | Lee, S, Rossmann, M.G. | | Deposit date: | 1996-03-22 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly.
Structure, 4, 1996
|
|
6NZ4
 
 | | YcjX-GDP (type I) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, YcjX Stress Protein | | Authors: | Lee, S, Tsai, J, Tsai, F.T. | | Deposit date: | 2019-02-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of the YcjX Stress Protein Reveals a Ras-Like GTP-Binding Protein.
J.Mol.Biol., 431, 2019
|
|
8JZI
 
 | | Mutant S-adenosylmethionine synthase from C. glutamicum | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Lee, S, Kim, K.J. | | Deposit date: | 2023-07-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and Biochemical Studies on Product Inhibition of S-Adenosylmethionine Synthetase from Corynebacterium glutamicum .
J.Agric.Food Chem., 71, 2023
|
|
8JZH
 
 | | C. glutamicum S-adenosylmethionine synthase | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, S-adenosylmethionine synthase, ... | | Authors: | Lee, S, Kim, K.J. | | Deposit date: | 2023-07-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Biochemical Studies on Product Inhibition of S-Adenosylmethionine Synthetase from Corynebacterium glutamicum .
J.Agric.Food Chem., 71, 2023
|
|
8JZG
 
 | | C. glutamicum S-adenosylmethionine synthase co-crystallized with Adenosine, triphosphate, and SAM | | Descriptor: | ADENOSINE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Lee, S, Kim, K.J. | | Deposit date: | 2023-07-05 | | Release date: | 2023-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural and Biochemical Studies on Product Inhibition of S-Adenosylmethionine Synthetase from Corynebacterium glutamicum .
J.Agric.Food Chem., 71, 2023
|
|
6N8V
 
 | | Hsp104DWB open conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 104 | | Authors: | Lee, S, Rho, S.H, Lee, J, Sung, N, Liu, J, Tsai, F.T.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Cryo-EM Structures of the Hsp104 Protein Disaggregase Captured in the ATP Conformation.
Cell Rep, 26, 2019
|
|
6N8Z
 
 | | HSP104DWB extended conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 104 | | Authors: | Lee, S, Rho, S.H, Lee, J, Sung, N, Liu, J, Tsai, F.T.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Cryo-EM Structures of the Hsp104 Protein Disaggregase Captured in the ATP Conformation.
Cell Rep, 26, 2019
|
|
6N8T
 
 | | Hsp104DWB closed conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 104 | | Authors: | Lee, S, Rho, S.H, Lee, J, Sung, N, Liu, J, Tsai, F.T.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Cryo-EM Structures of the Hsp104 Protein Disaggregase Captured in the ATP Conformation.
Cell Rep, 26, 2019
|
|
8K3F
 
 | |
1WYK
 
 | | SINDBIS VIRUS CAPSID PROTEIN (114-264) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, FORMYL GROUP, SINDBIS VIRUS CAPSID PROTEIN | | Authors: | Lee, S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 1998-01-12 | | Release date: | 1998-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the potential glycoprotein binding site of sindbis virus capsid protein with dioxane and model building.
Proteins, 33, 1998
|
|
8FDS
 
 | | Ankyrin domain of SKD3 isoform 2 | | Descriptor: | Caseinolytic peptidase B protein homolog, FORMIC ACID | | Authors: | Lee, S, Tsai, F.T.F, Chang, C. | | Deposit date: | 2022-12-04 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis of impaired disaggregase function in the oxidation-sensitive SKD3 mutant causing 3-methylglutaconic aciduria.
Nat Commun, 14, 2023
|
|
4JX7
 
 | | Crystal structure of Pim1 kinase in complex with inhibitor 2-[(trans-4-aminocyclohexyl)amino]-4-{[3-(trifluoromethyl)phenyl]amino}pyrido[4,3-d]pyrimidin-5(6H)-one | | Descriptor: | 2-[(trans-4-aminocyclohexyl)amino]-4-{[3-(trifluoromethyl)phenyl]amino}pyrido[4,3-d]pyrimidin-5(6H)-one, PIM1 consensus peptide, Serine/threonine-protein kinase pim-1 | | Authors: | Lee, S.J, Han, B.G, Cho, J.W, Choi, J.S, Lee, J.K, Song, H.J, Koh, J.S, Lee, B.I. | | Deposit date: | 2013-03-27 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of pim1 kinase in complex with a pyrido[4,3-d]pyrimidine derivative suggests a unique binding mode.
Plos One, 8, 2013
|
|
4JX3
 
 | | Crystal structure of Pim1 kinase | | Descriptor: | Serine/threonine-protein kinase pim-1 | | Authors: | Lee, S.J, Han, B.G, Cho, J.W, Choi, J.S, Lee, J.K, Song, H.J, Koh, J.S, Lee, B.I. | | Deposit date: | 2013-03-27 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of pim1 kinase in complex with a pyrido[4,3-d]pyrimidine derivative suggests a unique binding mode.
Plos One, 8, 2013
|
|
6PGQ
 
 | |
7YMO
 
 | |
5HS9
 
 | |
5HS7
 
 | |
