9ISP
 
 | | DUF2436 domain which is frequently found in virulence proteins from Porphyromonas gingivalis | | Descriptor: | Protease PrtH | | Authors: | Kim, B, Hwang, J, Do, H, Lee, J.H. | | Deposit date: | 2024-07-18 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | First crystal structure of the DUF2436 domain of virulence proteins from Porphyromonas gingivalis.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
8HG9
 
 | | Cytochrome P450 steroid hydroxylase (BaCYP106A6) from Bacillus species | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 steroid hydroxylase | | Authors: | Do, H, Lee, J.H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal Structure and Biochemical Analysis of a Cytochrome P450 Steroid Hydroxylase ( Ba CYP106A6) from Bacillus Species.
J Microbiol Biotechnol., 33, 2023
|
|
4UIP
 
 | | The complex structure of extracellular domain of EGFR with Repebody (rAC1). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EPIDERMAL GROWTH FACTOR RECEPTOR, ... | | Authors: | Kang, Y.J, Cha, Y.J, Cho, H.S, Lee, J.J, Kim, H.S. | | Deposit date: | 2015-03-31 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Enzymatic Prenylation and Oxime Ligation for the Synthesis of Stable and Homogeneous Protein-Drug Conjugates for Targeted Therapy.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1MKD
 
 | | crystal structure of PDE4D catalytic domain and zardaverine complex | | Descriptor: | 6-(4-DIFLUOROMETHOXY-3-METHOXY-PHENYL)-2H-PYRIDAZIN-3-ONE, MAGNESIUM ION, Phosphodiesterase 4D, ... | | Authors: | Lee, M.E, Markowitz, J, Lee, J.-O, Lee, H. | | Deposit date: | 2002-08-29 | | Release date: | 2003-03-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of phosphodiesterase 4D and inhibitor complex
FEBS LETT., 530, 2002
|
|
5AYX
 
 | | Crystal structure of Human Quinolinate Phosphoribosyltransferase | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase [carboxylating] | | Authors: | Kang, G.B, Kim, M.-K, Im, Y.J, Lee, J.H, Youn, H.-S, An, J.Y, Lee, J.-G, Fukuoka, S.-I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
3GWJ
 
 | | Crystal structure of Antheraea pernyi arylphorin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylphorin, FORMIC ACID, ... | | Authors: | Ryu, K.S, Lee, J.O, Kwon, T.H, Kim, S. | | Deposit date: | 2009-04-01 | | Release date: | 2009-05-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The presence of monoglucosylated N196-glycan is important for the structural stability of storage protein, arylphorin
Biochem.J., 421, 2009
|
|
6N8V
 
 | | Hsp104DWB open conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 104 | | Authors: | Lee, S, Rho, S.H, Lee, J, Sung, N, Liu, J, Tsai, F.T.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Cryo-EM Structures of the Hsp104 Protein Disaggregase Captured in the ATP Conformation.
Cell Rep, 26, 2019
|
|
6N8Z
 
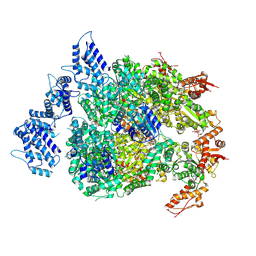 | | HSP104DWB extended conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 104 | | Authors: | Lee, S, Rho, S.H, Lee, J, Sung, N, Liu, J, Tsai, F.T.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Cryo-EM Structures of the Hsp104 Protein Disaggregase Captured in the ATP Conformation.
Cell Rep, 26, 2019
|
|
4ZTT
 
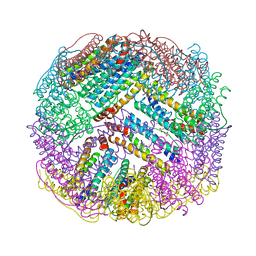 | | Crystal structures of ferritin mutants reveal diferric-peroxo intermediates | | Descriptor: | FE (II) ION, FE (III) ION, GLYCEROL, ... | | Authors: | Kim, S, Park, Y.H, Jung, S.W, Seok, J.H, Chung, Y.B, Lee, D.B, Gowda, G, Lee, J.H, Han, H.R, Cho, A.E, Lee, C, Chung, M.S, Kim, K.H. | | Deposit date: | 2015-05-15 | | Release date: | 2016-06-15 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis of Novel Iron-Uptake Route and Reaction Intermediates in Ferritins from Gram-Negative Bacteria.
J. Mol. Biol., 428, 2016
|
|
6NZ5
 
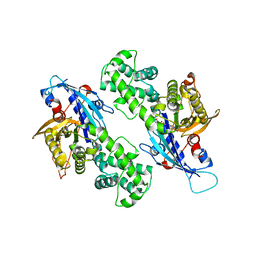 | | YcjX-GDPCP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, YcjX Stress Protein | | Authors: | Lee, S, Tsai, J, Tsai, F.T, Sung, N, Lee, J, Chang, C. | | Deposit date: | 2019-02-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | Crystal Structure of the YcjX Stress Protein Reveals a Ras-Like GTP-Binding Protein.
J.Mol.Biol., 431, 2019
|
|
6NZ6
 
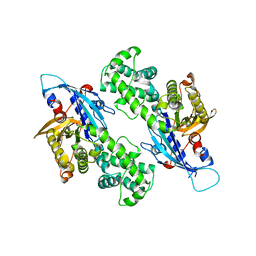 | | YcjX-GDP (type II) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, YcjX Stress Protein | | Authors: | Tsai, J, Sung, N, Lee, J, Chang, C, Lee, S, Tsai, F.T. | | Deposit date: | 2019-02-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the YcjX Stress Protein Reveals a Ras-Like GTP-Binding Protein.
J.Mol.Biol., 431, 2019
|
|
6N8T
 
 | | Hsp104DWB closed conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 104 | | Authors: | Lee, S, Rho, S.H, Lee, J, Sung, N, Liu, J, Tsai, F.T.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Cryo-EM Structures of the Hsp104 Protein Disaggregase Captured in the ATP Conformation.
Cell Rep, 26, 2019
|
|
1EW4
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI CYAY PROTEIN REVEALS A NOVEL FOLD FOR THE FRATAXIN FAMILY | | Descriptor: | CYAY PROTEIN | | Authors: | Suh, S.W, Cho, S, Lee, M.G, Yang, J.K, Lee, J.Y, Song, H.K. | | Deposit date: | 2000-04-22 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of Escherichia coli CyaY protein reveals a previously unidentified fold for the evolutionarily conserved frataxin family.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5CBN
 
 | | Fusion protein of mbp3-16 and B4 domain of protein A from staphylococcal aureus with chemical cross-linker EY-CBS | | Descriptor: | 2,2'-ethyne-1,2-diylbis{5-[(chloroacetyl)amino]benzenesulfonic acid}, Maltose-binding periplasmic protein, mbp3-16,Immunoglobulin G-binding protein A | | Authors: | Jeong, W.H, Lee, H, Song, D.H, Lee, J.O. | | Deposit date: | 2015-07-01 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Connecting two proteins using a fusion alpha helix stabilized by a chemical cross linker.
Nat Commun, 7, 2016
|
|
1J36
 
 | | Crystal Structure of Drosophila AnCE | | Descriptor: | ZINC ION, [N2-[(S)-1-CARBOXY-3-PHENYLPROPYL]-L-LYSYL-L-PROLINE, angiotensin converting enzyme | | Authors: | Kim, H.M, Shin, D.R, Yoo, O.J, Lee, H, Lee, J.-O. | | Deposit date: | 2003-01-20 | | Release date: | 2003-07-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Drosophila angiotensin I-converting enzyme bound to captopril and lisinopril
Febs Lett., 538, 2003
|
|
5CBO
 
 | | Fusion protein of mbp3-16 and B4 domain of protein A from staphylococcal aureus | | Descriptor: | mbp3-16,Immunoglobulin G-binding protein A | | Authors: | Jeong, W.H, Lee, H, Song, D.H, Lee, J.O. | | Deposit date: | 2015-07-01 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Connecting two proteins using a fusion alpha helix stabilized by a chemical cross linker.
Nat Commun, 7, 2016
|
|
5COC
 
 | | Fusion protein of human calmodulin and B4 domain of protein A from staphylococcal aureus | | Descriptor: | CALCIUM ION, Immunoglobulin G-binding protein A,Calmodulin | | Authors: | Jeong, W.H, Lee, H, Song, D.H, Lee, J.O. | | Deposit date: | 2015-07-20 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6691 Å) | | Cite: | Connecting two proteins using a fusion alpha helix stabilized by a chemical cross linker.
Nat Commun, 7, 2016
|
|
1J37
 
 | | Crystal Structure of Drosophila AnCE | | Descriptor: | L-CAPTOPRIL, ZINC ION, angiotensin converting enzyme | | Authors: | Kim, H.M, Shin, D.R, Yoo, O.J, Lee, H, Lee, J.-O. | | Deposit date: | 2003-01-20 | | Release date: | 2003-07-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Drosophila angiotensin I-converting enzyme bound to captopril and lisinopril
Febs Lett., 538, 2003
|
|
1J38
 
 | | Crystal Structure of Drosophila AnCE | | Descriptor: | ZINC ION, angiotensin converting enzyme | | Authors: | Kim, H.M, Shin, D.R, Lee, H, Lee, J.-O. | | Deposit date: | 2003-01-20 | | Release date: | 2003-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Drosophila angiotensin I-converting enzyme bound to captopril and lisinopril
Febs Lett., 538, 2003
|
|
4JX7
 
 | | Crystal structure of Pim1 kinase in complex with inhibitor 2-[(trans-4-aminocyclohexyl)amino]-4-{[3-(trifluoromethyl)phenyl]amino}pyrido[4,3-d]pyrimidin-5(6H)-one | | Descriptor: | 2-[(trans-4-aminocyclohexyl)amino]-4-{[3-(trifluoromethyl)phenyl]amino}pyrido[4,3-d]pyrimidin-5(6H)-one, PIM1 consensus peptide, Serine/threonine-protein kinase pim-1 | | Authors: | Lee, S.J, Han, B.G, Cho, J.W, Choi, J.S, Lee, J.K, Song, H.J, Koh, J.S, Lee, B.I. | | Deposit date: | 2013-03-27 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of pim1 kinase in complex with a pyrido[4,3-d]pyrimidine derivative suggests a unique binding mode.
Plos One, 8, 2013
|
|
4JX3
 
 | | Crystal structure of Pim1 kinase | | Descriptor: | Serine/threonine-protein kinase pim-1 | | Authors: | Lee, S.J, Han, B.G, Cho, J.W, Choi, J.S, Lee, J.K, Song, H.J, Koh, J.S, Lee, B.I. | | Deposit date: | 2013-03-27 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of pim1 kinase in complex with a pyrido[4,3-d]pyrimidine derivative suggests a unique binding mode.
Plos One, 8, 2013
|
|
8J26
 
 | | CryoEM structure of SARS CoV-2 RBD and Aptamer complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AM032-4, AM047-6, ... | | Authors: | Rahman, M.S, Jang, S.K, Lee, J.O. | | Deposit date: | 2023-04-14 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-Guided Development of Bivalent Aptamers Blocking SARS-CoV-2 Infection.
Molecules, 28, 2023
|
|
8J1Q
 
 | | CryoEM structure of SARS CoV-2 RBD and Aptamer complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AM032-0, AM047-0, ... | | Authors: | Rahman, M.S, Jang, S.K, Lee, J.O. | | Deposit date: | 2023-04-13 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-Guided Development of Bivalent Aptamers Blocking SARS-CoV-2 Infection.
Molecules, 28, 2023
|
|
1BGY
 
 | | CYTOCHROME BC1 COMPLEX FROM BOVINE | | Descriptor: | CYTOCHROME BC1 COMPLEX, FE2/S2 (INORGANIC) CLUSTER, HEME C, ... | | Authors: | Iwata, S, Lee, J.W, Okada, K, Lee, J.K, Iwata, M, Ramaswamy, S, Jap, B.K. | | Deposit date: | 1998-06-02 | | Release date: | 1999-01-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex.
Science, 281, 1998
|
|
5EWX
 
 | | Fusion protein of T4 lysozyme and B4 domain of protein A from staphylococcal aureus with chemical cross-linker EY-CBS | | Descriptor: | 2,2'-ethyne-1,2-diylbis{5-[(chloroacetyl)amino]benzenesulfonic acid}, Endolysin,Immunoglobulin G-binding protein A,Endolysin | | Authors: | Jeong, W.H, Lee, H, Song, D.H, Lee, J.O. | | Deposit date: | 2015-11-22 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Connecting two proteins using a fusion alpha helix stabilized by a chemical cross linker.
Nat Commun, 7, 2016
|
|
