2LXK
 
 | | Backbone 1H, 13C, and 15N Chemical Shift Assignments for cold shock protein, LmCsp | | Descriptor: | Cold shock-like protein CspLA | | Authors: | Lee, J, Jeong, K, Kim, Y. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Dynamic Features of Cold-Shock Proteins of Listeriamonocytogenes, a Psychrophilic Bacterium
Biochemistry, 52, 2013
|
|
7DOG
 
 | |
2PLK
 
 | |
2PYY
 
 | | Crystal Structure of the GluR0 ligand-binding core from Nostoc punctiforme in complex with (L)-glutamate | | Descriptor: | GLUTAMIC ACID, Ionotropic glutamate receptor bacterial homologue | | Authors: | Lee, J.H, Kang, G.B, Lim, H.-H, Ree, M, Park, C.-S, Eom, S.H. | | Deposit date: | 2007-05-17 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the GluR0 ligand-binding core from Nostoc punctiforme in complex with L-glutamate: structural dissection of the ligand interaction and subunit interface.
J.Mol.Biol., 376, 2008
|
|
2PLJ
 
 | |
7JP1
 
 | |
7JW5
 
 | | Crystal structure of WT-CYP199A4 in complex with 4-phenylbenzoic acid | | Descriptor: | CHLORIDE ION, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2020-08-24 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.526 Å) | | Cite: | Different Geometric Requirements for Cytochrome P450-Catalyzed Aliphatic Versus Aromatic Hydroxylation Results in Chemoselective Oxidation
Acs Catalysis, 12, 2022
|
|
7JOY
 
 | |
7KHP
 
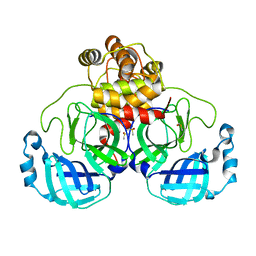 | | Acyl-enzyme intermediate structure of SARS-CoV-2 Mpro in complex with its C-terminal autoprocessing sequence. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Lee, J, Worrall, L.J, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2020-10-21 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site.
Nat Commun, 11, 2020
|
|
6IKX
 
 | |
6K8D
 
 | |
6KNL
 
 | |
6IKZ
 
 | |
6KNJ
 
 | |
6BQ1
 
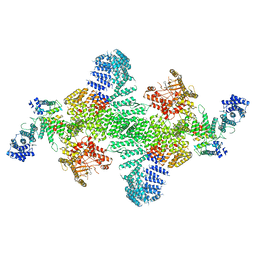 | | Human PI4KIIIa lipid kinase complex | | Descriptor: | 5-{2-amino-1-[4-(morpholin-4-yl)phenyl]-1H-benzimidazol-6-yl}-N-(2-fluorophenyl)-2-methoxypyridine-3-sulfonamide, Phosphatidylinositol 4-kinase III alpha (PI4KA), Protein FAM126A, ... | | Authors: | Lees, J.A, Zhang, Y, Oh, M, Schauder, C.M, Yu, X, Baskin, J, Dobbs, K, Notarangelo, L.D, Camilli, P.D, Walz, T, Reinisch, K.M. | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of the human PI4KIII alpha lipid kinase complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5TOD
 
 | | Transmembrane protein 24 SMP domain | | Descriptor: | Transmembrane protein 24 | | Authors: | Lees, J.A, Reinisch, K.M. | | Deposit date: | 2016-10-17 | | Release date: | 2017-03-01 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Lipid transport by TMEM24 at ER-plasma membrane contacts regulates pulsatile insulin secretion.
Science, 355, 2017
|
|
7UJA
 
 | | Cryo-EM structure of Human respiratory syncytial virus F variant (construct pXCS847A) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AM14 Fab heavy chain, AM14 Fab light chain, ... | | Authors: | Lees, J.A, Ammirati, M, Han, S. | | Deposit date: | 2022-03-30 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Rational design of a highly immunogenic prefusion-stabilized F glycoprotein antigen for a respiratory syncytial virus vaccine.
Sci Transl Med, 15, 2023
|
|
8TB7
 
 | | Cryo-EM Structure of GPR61- | | Descriptor: | 6-{[(3,5-difluoropyridin-4-yl)methyl]amino}-N-(4-ethoxy-6-methylpyrimidin-2-yl)-2-methoxy-N-(2-methoxyethyl)pyridine-3-sulfonamide, Fab hinge-binding nanobody, Fab24 BAK5 heavy chain, ... | | Authors: | Lees, J.A, Dias, J.M, Han, S. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | An inverse agonist of orphan receptor GPR61 acts by a G protein-competitive allosteric mechanism.
Nat Commun, 14, 2023
|
|
8TB0
 
 | | Cryo-EM Structure of GPR61-G protein complex stabilized by scFv16 | | Descriptor: | GPR61 fused to dominant negative G alpha S/I N18 chimera, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Lees, J.A, Dias, J.M, Han, S. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | An inverse agonist of orphan receptor GPR61 acts by a G protein-competitive allosteric mechanism.
Nat Commun, 14, 2023
|
|
7L7K
 
 | |
7L7F
 
 | |
7K1W
 
 | |
7K2V
 
 | |
7K1Y
 
 | |
8X3N
 
 | |
