5ACO
 
 | | Cryo-EM structure of PGT128 Fab in complex with BG505 SOSIP.664 Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 ENVELOPE GLYCOPROTEIN, ... | | Authors: | Lee, J.H, Ward, A.B. | | Deposit date: | 2015-08-17 | | Release date: | 2015-09-30 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Model Building and Refinement of a Natively Glycosylated HIV-1 Env Protein by High-Resolution Cryoelectron Microscopy.
Structure, 23, 2015
|
|
5AWN
 
 | |
3NE2
 
 | |
2AOQ
 
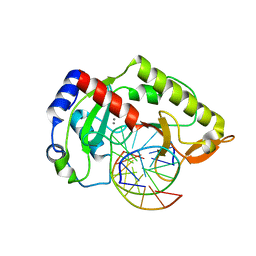 | | Crystal structure of MutH-unmethylated DNA complex | | Descriptor: | 5'-D(*GP*CP*AP*TP*GP*AP*TP*CP*AP*TP*GP*C)-3', CALCIUM ION, DNA mismatch repair protein mutH | | Authors: | Lee, J.Y, Chang, J, Joseph, N, Ghirlando, R, Rao, D.N, Yang, W. | | Deposit date: | 2005-08-13 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MutH complexed with hemi- and unmethylated DNAs: coupling base recognition and DNA cleavage.
Mol.Cell, 20, 2005
|
|
3HGQ
 
 | |
3HL4
 
 | | Crystal structure of a mammalian CTP:phosphocholine cytidylyltransferase with CDP-choline | | Descriptor: | Choline-phosphate cytidylyltransferase A, FORMIC ACID, GLYCEROL, ... | | Authors: | Lee, J, Paetzel, M, Cornell, R.B. | | Deposit date: | 2009-05-26 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a mammalian CTP: Phosphocholine cytidylyltransferase catalytic domain reveals novel active site residues within a highly conserved nucleotidyl-transferase fold
J.Biol.Chem., 284, 2009
|
|
2B02
 
 | | Crystal Structure of ARNT PAS-B Domain | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator | | Authors: | Lee, J, Botuyan, M.V, Nomine, Y, Ohh, M, Thompson, J.R, Mer, G. | | Deposit date: | 2005-09-12 | | Release date: | 2006-10-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure and Binding Properties of ARNT PAS-B Heterodimerization Domain
To be Published
|
|
1GUX
 
 | | RB POCKET BOUND TO E7 LXCXE MOTIF | | Descriptor: | ONCOPROTEIN, RETINOBLASTOMA PROTEIN | | Authors: | Lee, J.O, Russo, A.A, Pavletich, N.P. | | Deposit date: | 1997-11-15 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7.
Nature, 391, 1998
|
|
1PIB
 
 | | Solution structure of DNA containing CPD opposited by GA | | Descriptor: | 5'-D(*CP*GP*CP*AP*TP*TP*AP*CP*GP*C)-3', 5'-D(*GP*CP*GP*TP*GP*AP*TP*GP*CP*G)-3' | | Authors: | Lee, J.H, Park, C.J, Shin, J.S, Choi, B.S. | | Deposit date: | 2003-05-30 | | Release date: | 2004-05-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the DNA decamer duplex containing double T*G mismatches of cis-syn cyclobutane pyrimidine dimer: implications for DNA damage recognition by the XPC-hHR23B complex.
Nucleic Acids Res., 32, 2004
|
|
2EVU
 
 | | Crystal structure of aquaporin AqpM at 2.3A resolution | | Descriptor: | Aquaporin aqpM, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Lee, J.K, Kozono, D, Remis, J, Kitagawa, Y, Agre, P, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2005-10-31 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 A.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
5T42
 
 | | Structure of the Ebola virus envelope protein MPER/TM domain and its interaction with the fusion loop explains their fusion activity | | Descriptor: | Envelope glycoprotein | | Authors: | Lee, J, Nyenhuis, D.A, Nelson, E.A, Cafiso, D.S, White, J.M, Tamm, L.K. | | Deposit date: | 2016-08-28 | | Release date: | 2017-08-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ebola virus envelope protein MPER/TM domain and its interaction with the fusion loop explains their fusion activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3H2U
 
 | | Human raver1 RRM1, RRM2, and RRM3 domains in complex with human vinculin tail domain Vt | | Descriptor: | Raver-1, Vinculin | | Authors: | Lee, J.H, Rangarajan, E.S, Yogesha, S.D, Izard, T. | | Deposit date: | 2009-04-14 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Raver1 interactions with vinculin and RNA suggest a feed-forward pathway in directing mRNA to focal adhesions
Structure, 17, 2009
|
|
5V8M
 
 | | BG505 SOSIP.664 trimer in complex with broadly neutralizing HIV antibody 3BNC117 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, J.H, Cottrell, C.A, Ward, A.B. | | Deposit date: | 2017-03-22 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A Broadly Neutralizing Antibody Targets the Dynamic HIV Envelope Trimer Apex via a Long, Rigidified, and Anionic beta-Hairpin Structure.
Immunity, 46, 2017
|
|
1JYS
 
 | | Crystal Structure of E. coli MTA/AdoHcy Nucleosidase | | Descriptor: | ADENINE, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2001-09-13 | | Release date: | 2002-10-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of E. coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase reveals similarity to the purine nucleoside phosphorylases.
Structure, 9, 2001
|
|
5V8L
 
 | | BG505 SOSIP.664 trimer in complex with broadly neutralizing HIV antibodies 3BNC117 and PGT145 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3BNC117 antibody, ... | | Authors: | Lee, J.H, Ward, A.B. | | Deposit date: | 2017-03-22 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A Broadly Neutralizing Antibody Targets the Dynamic HIV Envelope Trimer Apex via a Long, Rigidified, and Anionic beta-Hairpin Structure.
Immunity, 46, 2017
|
|
5UMV
 
 | |
1MQV
 
 | | Crystal Structure of the Q1A/F32W/W72F mutant of Rhodopseudomonas palustris cytochrome c' (prime) expressed in E. coli | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Lee, J.C, Engman, K.C, Tezcan, F.A, Gray, H.B, Winkler, J.R. | | Deposit date: | 2002-09-17 | | Release date: | 2002-11-20 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Features of Cytochrome c' Folding Intermediates Revealed by Fluorescence Energy-Transfer Kinetics
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3N8X
 
 | | Crystal Structure of Cyclooxygenase-1 in Complex with Nimesulide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-NITRO-2-PHENOXYMETHANESULFONANILIDE, ... | | Authors: | Lee, J.Y. | | Deposit date: | 2010-05-28 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Comparison of Cyclooxygenase-1 Crystal Structures: Cross-Talk between Monomers Comprising Cyclooxygenase-1 Homodimers
Biochemistry, 49, 2010
|
|
2AQZ
 
 | | Crystal structure of FGF-1, S17T/N18T/G19 deletion mutant | | Descriptor: | Heparin-binding growth factor 1, SULFATE ION | | Authors: | Lee, J, Blaber, M. | | Deposit date: | 2005-08-18 | | Release date: | 2006-02-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conversion of type I 4:6 to 3:5 beta-turn types in human acidic fibroblast growth factor: Effects upon structure, stability, folding, and mitogenic function.
Proteins, 62, 2006
|
|
3O4A
 
 | |
3BAG
 
 | |
3BAQ
 
 | |
3BAU
 
 | |
3BAH
 
 | |
3BAO
 
 | |
