8G36
 
 | | Crystal structure of F182L-CYP199A4 in complex with terephthalic acid | | Descriptor: | CHLORIDE ION, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lee, J.H.Z, Bruning, J.B, Bell, S.G. | | Deposit date: | 2023-02-06 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering C-C Bond Cleavage Activity into a P450 Monooxygenase Enzyme.
J.Am.Chem.Soc., 145, 2023
|
|
8G35
 
 | | Crystal structure of F182L-CYP199A4 in complex with (S)-4-(2-hydroxy-3-oxobutan-2-yl)benzoic acid | | Descriptor: | 4-[(2S)-2-hydroxy-3-oxobutan-2-yl]benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Lee, J.H.Z, Bell, S.G, Bruning, J.B. | | Deposit date: | 2023-02-06 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering C-C Bond Cleavage Activity into a P450 Monooxygenase Enzyme.
J.Am.Chem.Soc., 145, 2023
|
|
9J8E
 
 | | Structural insights into BirA from Haemophilus influenzae, a bifunctional protein as a biotin protein ligase and a transcriptional repressor | | Descriptor: | BIOTINYL-5-AMP, Bifunctional ligase/repressor BirA | | Authors: | Lee, J.Y, Jeong, K.H, Son, S.B, Ko, J.H. | | Deposit date: | 2024-08-21 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into BirA from Haemophilus influenzae, a bifunctional protein as a biotin protein ligase and a transcriptional repressor.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
9J8F
 
 | | Structural insights into BirA from Haemophilus influenzae, a bifunctional protein as a biotin protein ligase and a transcriptional repressor | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Bifunctional ligase/repressor BirA, PENTAETHYLENE GLYCOL | | Authors: | Lee, J.Y, Jeong, K.H, Son, S.B, Ko, J.H. | | Deposit date: | 2024-08-21 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into BirA from Haemophilus influenzae, a bifunctional protein as a biotin protein ligase and a transcriptional repressor.
Biochem.Biophys.Res.Commun., 733, 2024
|
|
1Z5O
 
 | | Crystal structure of MTA/AdoHcy nucleosidase Asp197Asn mutant complexed with 5'-methylthioadenosine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Smith, G.D, Horvatin, C, Huang, D.J.T, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2005-03-18 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis
J.Mol.Biol., 352, 2005
|
|
1Z5N
 
 | | Crystal structure of MTA/AdoHcy nucleosidase Glu12Gln mutant complexed with 5-methylthioribose and adenine | | Descriptor: | 5-S-methyl-5-thio-alpha-D-ribofuranose, ADENINE, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Smith, G.D, Horvatin, C, Huang, D.J.T, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2005-03-18 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis
J.Mol.Biol., 352, 2005
|
|
5H3H
 
 | | Esterase (EaEST) from Exiguobacterium antarcticum | | Descriptor: | Abhydrolase domain-containing protein, ETHANEPEROXOIC ACID | | Authors: | Lee, J.H, Lee, C.W. | | Deposit date: | 2016-10-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure and Functional Characterization of an Esterase (EaEST) from Exiguobacterium antarcticum.
Plos One, 12, 2017
|
|
4MRN
 
 | | Structure of a bacterial Atm1-family ABC transporter | | Descriptor: | ABC transporter related protein, LAURYL DIMETHYLAMINE-N-OXIDE, PHOSPHATE ION | | Authors: | Lee, J.Y, Yang, J.G, Zhitnitsky, D, Lewinson, O, Rees, D.C. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for heavy metal detoxification by an Atm1-type ABC exporter.
Science, 343, 2014
|
|
4MRS
 
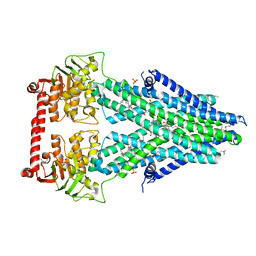 | | Structure of a bacterial Atm1-family ABC transporter | | Descriptor: | ABC transporter related protein, LAURYL DIMETHYLAMINE-N-OXIDE, OXIDIZED GLUTATHIONE DISULFIDE, ... | | Authors: | Lee, J.Y, Yang, J.G, Zhitnitsky, D, Lewinson, O, Rees, D.C. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for heavy metal detoxification by an Atm1-type ABC exporter.
Science, 343, 2014
|
|
4MRR
 
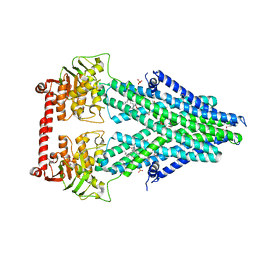 | | Structure of a bacterial Atm1-family ABC transporter | | Descriptor: | ABC transporter related protein, LAURYL DIMETHYLAMINE-N-OXIDE, PHOSPHATE ION, ... | | Authors: | Lee, J.Y, Yang, J.G, Zhitnitsky, D, Lewinson, O, Rees, D.C. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural basis for heavy metal detoxification by an Atm1-type ABC exporter.
Science, 343, 2014
|
|
5XWB
 
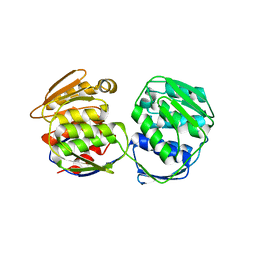 | | Crystal Structure of 5-Enolpyruvulshikimate-3-phosphate Synthase from a Psychrophilic Bacterium, Colwellia psychrerythraea | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase | | Authors: | Lee, J.H, Kim, H.J, Choi, J.M, Kim, D.-W, Seo, Y.-S. | | Deposit date: | 2017-06-29 | | Release date: | 2017-09-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of 5-enolpyruvylshikimate-3-phosphate synthase from a psychrophilic bacterium, Colwellia psychrerythraea 34H.
Biochem. Biophys. Res. Commun., 492, 2017
|
|
4MRV
 
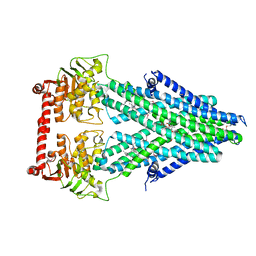 | | Structure of a bacterial Atm1-family ABC transporter | | Descriptor: | ABC transporter related protein, LAURYL DIMETHYLAMINE-N-OXIDE, PHOSPHATE ION, ... | | Authors: | Lee, J.Y, Yang, J.G, Zhitnitsky, D, Lewinson, O, Rees, D.C. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for heavy metal detoxification by an Atm1-type ABC exporter.
Science, 343, 2014
|
|
4MVD
 
 | | Crystal Structure of a Mammalian Cytidylyltransferase | | Descriptor: | Choline-phosphate cytidylyltransferase A, [2-CYTIDYLATE-O'-PHOSPHONYLOXYL]-ETHYL-TRIMETHYL-AMMONIUM | | Authors: | Lee, J, Cornell, R.B. | | Deposit date: | 2013-09-23 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (8 Å) | | Cite: | Structural Basis for Autoinhibition of CTP:Phosphocholine Cytidylyltransferase (CCT), the Regulatory Enzyme in Phosphatidylcholine Synthesis, by Its Membrane-binding Amphipathic Helix.
J.Biol.Chem., 289, 2014
|
|
9KOK
 
 | | Crystal structure of ExaC, an NAD+-dependent aldehyde dehydrogenase, from Pseudomonas aeruginosa | | Descriptor: | GLYCEROL, MAGNESIUM ION, NAD+ dependent aldehyde dehydrogenase ExaC, ... | | Authors: | Lee, J.Y, Ko, J.H, Jeong, K.H, Son, S.B. | | Deposit date: | 2024-11-20 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of ExaC, an NAD + -dependent aldehyde dehydrogenase, from Pseudomonas aeruginosa.
Biochem.Biophys.Res.Commun., 742, 2025
|
|
9KOI
 
 | | Crystal structure of ExaC, an NAD+-dependent aldehyde dehydrogenase, from Pseudomonas aeruginosa | | Descriptor: | NAD+ dependent aldehyde dehydrogenase ExaC | | Authors: | Lee, J.Y, Ko, J.H, Jeong, K.H, Son, S.B. | | Deposit date: | 2024-11-20 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of ExaC, an NAD + -dependent aldehyde dehydrogenase, from Pseudomonas aeruginosa.
Biochem.Biophys.Res.Commun., 742, 2025
|
|
5GPA
 
 | |
6Z6P
 
 | | HDAC-PC-Nuc | | Descriptor: | DNA (145-MER), HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
8RZ9
 
 | | MutSbeta-ATPgS with kinked MSH2 clamp | | Descriptor: | DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, MAGNESIUM ION, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2024-02-12 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | MutSbeta-ATPgS complex with kinked MSH2 clamp
To Be Published
|
|
8RZ7
 
 | | (CAG)2 DNA-bound MutSbeta in open form with kinked MSH2 clamp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2024-02-12 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | (CAG)2 DNA-bound MutSbeta in open form with kinked MSH2 clamp
To Be Published
|
|
8RZ8
 
 | | MutSbeta-ATPgS with straight MSH2 clamp | | Descriptor: | DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, MAGNESIUM ION, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2024-02-12 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | MutSbeta-ATPgS complex with straight MSH2 clamp
To Be Published
|
|
7COE
 
 | | Crystal structure of Receptor binding domain of MERS-CoV and KNIH90-F1 Fab complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, ... | | Authors: | Lee, J.Y, Song, J.Y, Lee, H.S, Hong, E, Jang, T.H. | | Deposit date: | 2020-08-04 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of a novel antibody against the spike protein inhibits Middle East respiratory syndrome coronavirus infections.
Sci Rep, 12, 2022
|
|
7CV2
 
 | | Crystal structure of B. halodurans NiaR in niacin-bound form | | Descriptor: | NICOTINIC ACID, Transcriptional regulator NiaR, ZINC ION | | Authors: | Lee, J.Y, Lee, D.W, Park, Y.W, Lee, M.Y, Jeong, K.H. | | Deposit date: | 2020-08-25 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structural analysis and insight into effector binding of the niacin-responsive repressor NiaR from Bacillus halodurans.
Sci Rep, 10, 2020
|
|
6Z6H
 
 | | HDAC-DC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (8.55 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
6Z6O
 
 | | HDAC-TC | | Descriptor: | HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, Histone deacetylase HDA1, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
7CV0
 
 | | Crystal structure of B. halodurans NiaR in apo form | | Descriptor: | Transcriptional regulator NiaR, ZINC ION | | Authors: | Lee, J.Y, Lee, D.W, Park, Y.W, Lee, M.Y, Jeong, K.H. | | Deposit date: | 2020-08-25 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural analysis and insight into effector binding of the niacin-responsive repressor NiaR from Bacillus halodurans.
Sci Rep, 10, 2020
|
|
