8GUW
 
 | |
8KIC
 
 | | Bacterial serine protease | | Descriptor: | Lysozyme fragment (unknown sequence), peptidase Do | | Authors: | Lee, I.-G, Jeon, H. | | Deposit date: | 2023-08-23 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of bacterial serine protease
To Be Published
|
|
4ZG0
 
 | | Crystal structure of Mouse Syndesmos protein | | Descriptor: | Protein syndesmos | | Authors: | Lee, I, Kim, H, Yoo, J, Cho, H, Lee, W. | | Deposit date: | 2015-04-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of syndesmos and its interaction with Syndecan-4 proteoglycan
Biochem.Biophys.Res.Commun., 463, 2015
|
|
8W69
 
 | | DegQ-b-casein complex | | Descriptor: | Casein fragment, peptidase Do | | Authors: | Lee, I.-G, Jeon, H. | | Deposit date: | 2023-08-28 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of E. coli Deg-b-casein complex
To Be Published
|
|
5G1D
 
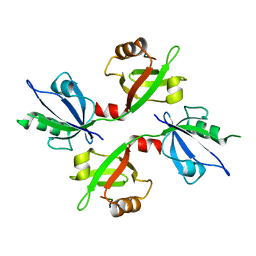 | | The complex structure of syntenin-1 PDZ domain with c-terminal extension | | Descriptor: | SYNDECAN-4, SYNTENIN-1 | | Authors: | Lee, I, Kim, H, Yun, J.H, Lee, W. | | Deposit date: | 2016-03-25 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | New Structural Insight of C-Terminal Region of Syntenin-1, Enhancing the Molecular Dimerization and Inhibitory Function Related on Syndecan-4 Signaling.
Sci.Rep., 6, 2016
|
|
5G1E
 
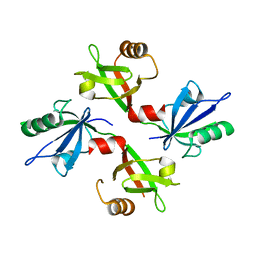 | | The complex structure of syntenin-1 PDZ domain with c-terminal extension | | Descriptor: | SYNTENIN-1 | | Authors: | Lee, I, Kim, H, Yun, J.H, Lee, W. | | Deposit date: | 2016-03-25 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | New Structural Insight of C-Terminal Region of Syntenin-1, Enhancing the Molecular Dimerization and Inhibitory Function Related on Syndecan-4 Signaling.
Sci.Rep., 6, 2016
|
|
5JSD
 
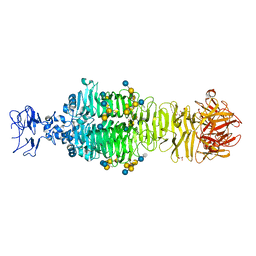 | | Crystal structure of phiAB6 tailspike in complex with five-repeated oligosaccharides of Acinetobacter baumannii surface polysaccharide | | Descriptor: | ACETIC ACID, MALONIC ACID, beta-D-galactopyranose-(1-3)-2-amino-2-deoxy-beta-D-galactopyranose-(1-3)-[5,7-bisacetamido-3,5,7,9-tetradeoxy-L-glycero-alpha-L-manno-non-2-ulopyranosonic acid-(2-6)-beta-D-glucopyranose-(1-6)]beta-D-galactopyranose-(1-3)-2-amino-2-deoxy-beta-D-galactopyranose-(1-3)-[beta-D-glucopyranose-(1-6)]beta-D-galactopyranose, ... | | Authors: | Lee, I.M, Tu, I.F, Huang, K.F, Wu, S.H. | | Deposit date: | 2016-05-08 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural basis for fragmenting the exopolysaccharide of Acinetobacter baumannii by bacteriophage Phi AB6 tailspike protein
Sci Rep, 7, 2017
|
|
5JS4
 
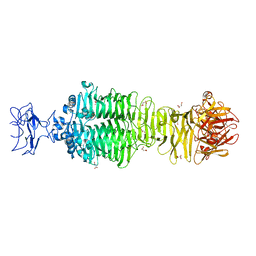 | | Crystal structure of phiAB6 tailspike | | Descriptor: | MALONIC ACID, phiAB6 tailspike | | Authors: | Lee, I.M, Tu, I.F, Huang, K.F, Wu, S.H. | | Deposit date: | 2016-05-07 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural basis for fragmenting the exopolysaccharide of Acinetobacter baumannii by bacteriophage Phi AB6 tailspike protein
Sci Rep, 7, 2017
|
|
5JSE
 
 | | Crystal structure of phiAB6 tailspike in complex with three-repeated oligosaccharides of Acinetobacter baumannii surface polysaccharide | | Descriptor: | ACETIC ACID, MALONIC ACID, beta-D-galactopyranose-(1-3)-2-amino-2-deoxy-beta-D-galactopyranose-(1-3)-[5,7-bisacetamido-3,5,7,9-tetradeoxy-L-glycero-alpha-L-manno-non-2-ulopyranosonic acid-(2-6)-beta-D-glucopyranose-(1-6)]beta-D-galactopyranose-(1-3)-2-amino-2-deoxy-beta-D-galactopyranose-(1-3)-[beta-D-glucopyranose-(1-6)]beta-D-galactopyranose, ... | | Authors: | Lee, I.M, Tu, I.F, Huang, K.F, Wu, S.H. | | Deposit date: | 2016-05-08 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for fragmenting the exopolysaccharide of Acinetobacter baumannii by bacteriophage Phi AB6 tailspike protein
Sci Rep, 7, 2017
|
|
3CMM
 
 | | Crystal Structure of the Uba1-Ubiquitin Complex | | Descriptor: | PROLINE, Ubiquitin, Ubiquitin-activating enzyme E1 1 | | Authors: | Lee, I, Schindelin, H. | | Deposit date: | 2008-03-23 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes.
Cell(Cambridge,Mass.), 134, 2008
|
|
6B9H
 
 | |
2YIJ
 
 | | Crystal Structure of phospholipase A1 | | Descriptor: | PHOSPHOLIPASE A1-IIGAMMA | | Authors: | Lee, I. | | Deposit date: | 2011-05-13 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Phospholipase A1
To be Published
|
|
6JF8
 
 | |
6JF4
 
 | |
6JFF
 
 | |
6JFA
 
 | |
6JF5
 
 | |
6JFE
 
 | |
6JF9
 
 | |
6JF3
 
 | |
6JFC
 
 | |
6JFN
 
 | |
6JFD
 
 | |
6JEV
 
 | |
6IKT
 
 | |
