2FTU
 
 | | solution structure of domain 3 of RAP | | Descriptor: | Alpha-2-macroglobulin receptor-associated protein, domain 3 | | Authors: | Lee, D, Walsh, J.D, Wang, Y.-X. | | Deposit date: | 2006-01-24 | | Release date: | 2006-05-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | RAP uses a histidine switch to regulate its interaction with LRP in the ER and Golgi.
Mol.Cell, 22, 2006
|
|
3DSI
 
 | | Crystal Structure of Arabidopsis thaliana Allene Oxide Synthase (AOS, cytochrome P450 74A, CYP74A) Complexed with 13(S)-HOT at 1.60 A resolution | | Descriptor: | (9Z,11E,13S,15Z)-13-hydroxyoctadeca-9,11,15-trienoic acid, Cytochrome P450 74A, chloroplast, ... | | Authors: | Lee, D.S, Nioche, P, Raman, C.S. | | Deposit date: | 2008-07-12 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes
Nature, 455, 2008
|
|
2AEX
 
 | | The 1.58A Crystal Structure of Human Coproporphyrinogen Oxidase Reveals the Structural Basis of Hereditary Coproporphyria | | Descriptor: | CITRIC ACID, Coproporphyrinogen III oxidase, mitochondrial | | Authors: | Lee, D.S, Flachsova, E, Bodnarova, M, Demeler, B, Martasek, P, Raman, C.S. | | Deposit date: | 2005-07-24 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural basis of hereditary coproporphyria.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1LS8
 
 | | NMR structure of the unliganded Bombyx mori pheromone-binding protein at physiological pH | | Descriptor: | pheromone binding protein | | Authors: | Lee, D, Damberger, F, Horst, R, Guntert, P, Leal, W.S, Wuthrich, K. | | Deposit date: | 2002-05-17 | | Release date: | 2002-11-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the unliganded Bombyx mori pheromone-binding protein at physiological pH
FEBS Lett., 531, 2002
|
|
6JX2
 
 | | Crystal structure of Ketol-acid reductoisomerase from Corynebacterium glutamicum | | Descriptor: | 1,2-ETHANEDIOL, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, ... | | Authors: | Lee, D, Hong, J, Kim, K.-J. | | Deposit date: | 2019-04-22 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and Biochemical Characterization of Ketol-Acid Reductoisomerase fromCorynebacterium glutamicum.
J.Agric.Food Chem., 67, 2019
|
|
2H8W
 
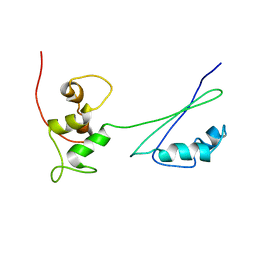 | | Solution structure of ribosomal protein L11 | | Descriptor: | 50S ribosomal protein L11 | | Authors: | Lee, D, Walsh, J.D, Yu, P, Choli-Papadopoulou, T, Krueger, S, Draper, D, Wang, Y.-X. | | Deposit date: | 2006-06-08 | | Release date: | 2007-02-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of Free L11 and Functional Dynamics of L11 in Free, L11-rRNA(58 nt) Binary and L11-rRNA(58 nt)-thiostrepton Ternary Complexes.
J.Mol.Biol., 367, 2007
|
|
1CQ0
 
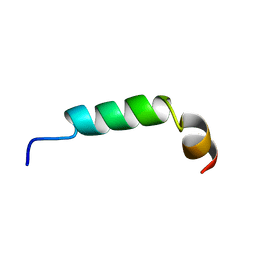 | | SOLUTION STRUCTURE OF A HUMAN HYPOCRETIN-2/OREXIN-B'SOLUTION STRUCTURE OF A HUMAN HYPOCRETIN-2/OREXIN-B ' | | Descriptor: | PROTEIN (NEW HYPOTHALAMIC NEUROPEPTIDE/OREXIN-B28) | | Authors: | Lee, K.-H, Bang, E.J, Chae, K.-J, Lee, D.W, Lee, W. | | Deposit date: | 1999-08-04 | | Release date: | 2000-01-10 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a new hypothalamic neuropeptide, human hypocretin-2/orexin-B.
Eur.J.Biochem., 266, 1999
|
|
1BXB
 
 | | XYLOSE ISOMERASE FROM THERMUS THERMOPHILUS | | Descriptor: | XYLOSE ISOMERASE | | Authors: | Chang, C, Park, B.C, Lee, D.-S, Suh, S.W. | | Deposit date: | 1998-10-02 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of thermostable xylose isomerases from Thermus caldophilus and Thermus thermophilus: possible structural determinants of thermostability.
J.Mol.Biol., 288, 1999
|
|
1BXC
 
 | | XYLOSE ISOMERASE FROM THERMUS CALDOPHILUS | | Descriptor: | XYLOSE ISOMERASE | | Authors: | Chang, C, Park, B.C, Lee, D.-S, Suh, S.W. | | Deposit date: | 1998-10-02 | | Release date: | 1999-02-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of thermostable xylose isomerases from Thermus caldophilus and Thermus thermophilus: possible structural determinants of thermostability.
J.Mol.Biol., 288, 1999
|
|
4UN2
 
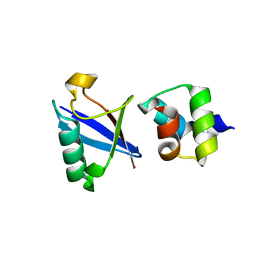 | | Crystal structure of the UBA domain of Dsk2 in complex with Ubiquitin | | Descriptor: | UBIQUITIN, UBIQUITIN DOMAIN-CONTAINING PROTEIN DSK2 | | Authors: | Michielssens, S, Peters, J.H, Ban, D, Pratihar, S, Seeliger, D, Sharma, M, Giller, K, Sabo, T.M, Becker, S, Lee, D, Griesinger, C, de Groot, B.L. | | Deposit date: | 2014-05-23 | | Release date: | 2014-08-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A Designed Conformational Shift to Control Protein Binding Specificity.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6A6E
 
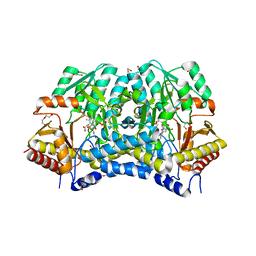 | | Crystal structure of thermostable Cysteine desulfurase (FiSufS) from thermophilic Fervidobacterium Islandicum AW-1 | | Descriptor: | CITRIC ACID, Cysteine desulfurase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dhanasingh, I, Jin, H.S, Lee, D.W, Lee, S.H. | | Deposit date: | 2018-06-27 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The sulfur formation system mediating extracellular cysteine-cystine recycling in Fervidobacterium islandicum AW-1 is associated with keratin degradation.
Microb Biotechnol, 2020
|
|
6A6F
 
 | | Crystal structure of Putative iron-sulfur cluster assembly scaffold protein for SUF system (FiSufU) from thermophilic Fervidobacterium Islandicum AW-1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Iron-sulfur cluster assembly scaffold protein NifU, ... | | Authors: | Dhanasingh, I, Jin, H.S, Lee, D.W, Lee, S.H. | | Deposit date: | 2018-06-27 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The sulfur formation system mediating extracellular cysteine-cystine recycling in Fervidobacterium islandicum AW-1 is associated with keratin degradation.
Microb Biotechnol, 2020
|
|
6A6G
 
 | | Crystal structure of thermostable FiSufS-SufU complex from thermophilic Fervidobacterium Islandicum AW-1 | | Descriptor: | Cysteine desulfurase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Dhanasingh, I, Jin, H.S, Lee, D.W, Lee, S.H. | | Deposit date: | 2018-06-27 | | Release date: | 2019-10-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The sulfur formation system mediating extracellular cysteine-cystine recycling in Fervidobacterium islandicum AW-1 is associated with keratin degradation.
Microb Biotechnol, 2020
|
|
1EJQ
 
 | | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN IN THE PRESENCE OF PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE | | Descriptor: | SYNDECAN-4 | | Authors: | Shin, J, Oh, E.S, Lee, D, Couchman, J.R, Lee, W. | | Deposit date: | 2000-03-04 | | Release date: | 2001-03-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN IN THE PRESENCE OF PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE
To be Published
|
|
1SGW
 
 | | Putative ABC transporter (ATP-binding protein) from Pyrococcus furiosus Pfu-867808-001 | | Descriptor: | CHLORIDE ION, SODIUM ION, putative ABC transporter | | Authors: | Liu, Z.J, Tempel, W, Shah, A, Chen, L, Lee, D, Kelley, L.-L.C, Dillard, B.D, Rose, J.P, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.S, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-02-24 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Putative ABC transporter (ATP-binding protein) from Pyrococcus furiosus Pfu-867808-001
To be Published
|
|
4ZRM
 
 | |
4ZRN
 
 | |
5B7Y
 
 | | Crystal Structure of Hyperthermophilic Thermotoga maritima L-Ketose-3-Epimerase with Co2+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, COBALT (II) ION, ... | | Authors: | Cao, T.P, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2016-06-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | TM0416, a Hyperthermophilic Promiscuous Nonphosphorylated Sugar Isomerase, Catalyzes Various C5and C6Epimerization Reactions
Appl. Environ. Microbiol., 83, 2017
|
|
5B7Z
 
 | | Crystal Structure of Hyperthermophilic Thermotoga maritima L-Ketose-3-Epimerase with Ni2+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, NICKEL (II) ION, ... | | Authors: | Cao, T.P, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2016-06-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | TM0416, a Hyperthermophilic Promiscuous Nonphosphorylated Sugar Isomerase, Catalyzes Various C5and C6Epimerization Reactions
Appl. Environ. Microbiol., 83, 2017
|
|
5B80
 
 | | Crystal Structure of Hyperthermophilic Thermotoga maritima L-Ketose-3-Epimerase with Cu2+ | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, COPPER (II) ION, Uncharacterized protein TM_0416 | | Authors: | Cao, T.P, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2016-06-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | TM0416, a Hyperthermophilic Promiscuous Nonphosphorylated Sugar Isomerase, Catalyzes Various C5and C6Epimerization Reactions
Appl. Environ. Microbiol., 83, 2017
|
|
8H24
 
 | | Leucine-rich alpha-2-glycoprotein 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Leucine-rich alpha-2-glycoprotein, SULFATE ION | | Authors: | Won, S.Y, Park, B.S, Lee, D.S, Kim, H.M, Han, A, Yang, J. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of LRG1 and the functional significance of LRG1 glycan for LPHN2 activation.
Exp.Mol.Med., 55, 2023
|
|
6WYV
 
 | | E. coli 50S ribosome bound to compounds 47 and VS1 | | Descriptor: | (3R,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2020-05-13 | | Release date: | 2020-06-17 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
7S1G
 
 | | wild-type Escherichia coli stalled ribosome with antibiotic linezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S1H
 
 | | Wild-type Escherichia coli ribosome with antibiotic linezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6PC5
 
 | | E. coli 50S ribosome bound to compounds 46 and VS1 | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-4,12-dimethyl-1,7,22-trioxo-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl isoquinolin-3-ylcarbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-16 | | Release date: | 2020-06-17 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
