3G0C
 
 | | Crystal structure of dipeptidyl peptidase IV in complex with a pyrimidinedione inhibitor 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(2-chlorobenzyl)-1,3-dimethyl-8-piperazin-1-yl-3,7-dihydro-1H-purine-2,6-dione, ... | | Authors: | Zhang, Z, Wallace, M.B, Feng, J, Stafford, J.A, Kaldor, S.W, Shi, L, Skene, R.J, Aertgeerts, K, Lee, B, Jennings, A, Xu, R, Kassel, D, Webb, D.R, Gwaltney, S.L. | | Deposit date: | 2009-01-27 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design and Synthesis of Pyrimidinone and Pyrimidinedione Inhibitors of Dipeptidyl Peptidase IV.
J.Med.Chem., 54, 2011
|
|
1NP3
 
 | | Crystal structure of class I acetohydroxy acid isomeroreductase from Pseudomonas aeruginosa | | Descriptor: | Ketol-acid reductoisomerase | | Authors: | Ahn, H.J, Eom, S.J, Yoon, H.-J, Lee, B.I, Cho, H, Suh, S.W. | | Deposit date: | 2003-01-17 | | Release date: | 2003-05-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Class I Acetohydroxy Acid Isomeroreductase from Pseudomonas aeruginosa
J.Mol.Biol., 328, 2003
|
|
6JHV
 
 | | Structure of anti-CRISPR AcrIIC3 | | Descriptor: | AcrIIC3 | | Authors: | Suh, J.Y, Lee, B.J, Lee, S.J, Kim, Y. | | Deposit date: | 2019-02-19 | | Release date: | 2019-08-28 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.321 Å) | | Cite: | Anti-CRISPR AcrIIC3 discriminates between Cas9 orthologs via targeting the variable surface of the HNH nuclease domain.
Febs J., 286, 2019
|
|
3LY6
 
 | |
2B9L
 
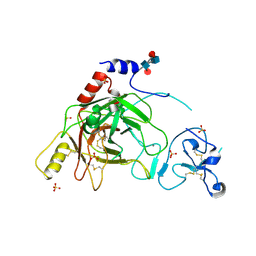 | | Crystal structure of prophenoloxidase activating factor-II from the beetle Holotrichia diomphalia | | Descriptor: | CALCIUM ION, SULFATE ION, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Piao, S, Song, Y.-L, Park, S.Y, Lee, B.L, Oh, B.-H, Ha, N.-C. | | Deposit date: | 2005-10-12 | | Release date: | 2006-01-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a clip-domain serine protease and functional roles of the clip domains.
Embo J., 24, 2005
|
|
2OTR
 
 | |
5B62
 
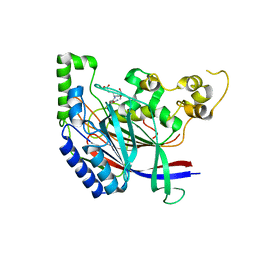 | | Crystal structure of N-terminal amidase with Asn-Glu-Ala peptide | | Descriptor: | ASN-GLU-ALA, Nta1p | | Authors: | Kim, M.K, Oh, S.-J, Lee, B.-G, Song, H.K. | | Deposit date: | 2016-05-24 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.042 Å) | | Cite: | Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5YU2
 
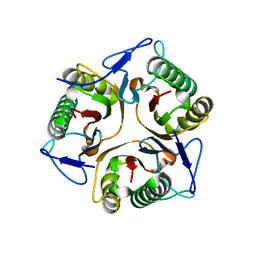 | | Structure of Ribonuclease YabJ | | Descriptor: | Translation initiation inhibitor homologue | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2017-11-20 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A novel chlorination-induced ribonuclease YabJ fromStaphylococcus aureus.
Biosci. Rep., 38, 2018
|
|
2DS5
 
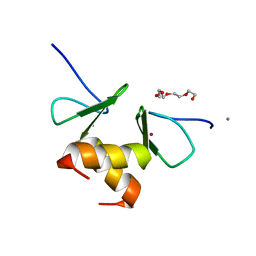 | | Structure of the ZBD in the orthorhomibic crystal from | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, CALCIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Song, H.K, Park, E.Y, Lee, B.G, Hong, S.B. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
1UAK
 
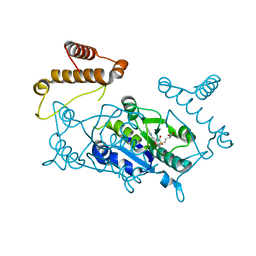 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
1UAL
 
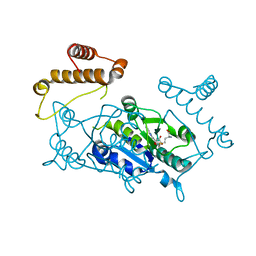 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
1UM0
 
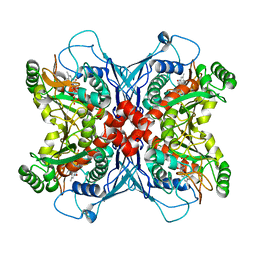 | | Crystal structure of chorismate synthase complexed with FMN | | Descriptor: | Chorismate synthase, FLAVIN MONONUCLEOTIDE | | Authors: | Ahn, H.J, Yoon, H.J, Lee, B, Suh, S.W. | | Deposit date: | 2003-09-18 | | Release date: | 2004-06-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of chorismate synthase: a novel FMN-binding protein fold and functional insights
J.Mol.Biol., 336, 2004
|
|
2DS6
 
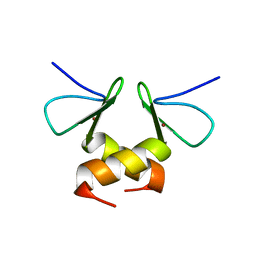 | | Structure of the ZBD in the tetragonal crystal form | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, ZINC ION | | Authors: | Park, E.Y, Lee, B.G, Hong, S.B, Song, H.K. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
2DS8
 
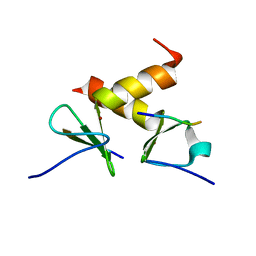 | | Structure of the ZBD-XB complex | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, SspB-tail peptide, ZINC ION | | Authors: | Park, E.Y, Lee, B.G, Hong, S.B, Kim, H.W, Song, H.K. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
1Z8M
 
 | |
1UMF
 
 | |
2DS7
 
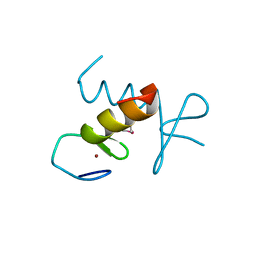 | | Structure of the ZBD in the hexagonal crystal form | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit clpX, ZINC ION | | Authors: | Park, E.Y, Lee, B.G, Hong, S.B, Song, H.K. | | Deposit date: | 2006-06-22 | | Release date: | 2007-02-13 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of SspB-tail Recognition by the Zinc Binding Domain of ClpX.
J.Mol.Biol., 367, 2007
|
|
5HYY
 
 | |
4Y1G
 
 | | SAV1875-E17N | | Descriptor: | Uncharacterized protein SAV1875 | | Authors: | Kim, H.J, Kwon, A.R, Lee, B.J. | | Deposit date: | 2015-02-07 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insight into the different oxidation states of SAV1875 from Staphylococcus aureus
Biochem.J., 473, 2016
|
|
4LS4
 
 | | Crystal structure of L66S mutant toxin from Helicobacter pylori | | Descriptor: | BROMIDE ION, Uncharacterized protein, Toxin | | Authors: | Pathak, C.C, Im, H, Lee, B.J, Yoon, H.J. | | Deposit date: | 2013-07-22 | | Release date: | 2014-02-05 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of apo and copper bound HP0894 toxin from Helicobacter pylori 26695 and insight into mRNase activity
Biochim.Biophys.Acta, 1834, 2013
|
|
7CQE
 
 | |
7R22
 
 | |
2PX9
 
 | | The intrinsic affinity between E2 and the Cys domain of E1 in Ubiquitin-like modifications | | Descriptor: | SUMO-activating enzyme subunit 2, SUMO-conjugating enzyme UBC9 | | Authors: | Wang, J.H, Hu, W.D, Cai, S, Lee, B, Song, J, Chen, Y. | | Deposit date: | 2007-05-14 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The intrinsic affinity between E2 and the Cys domain of E1 in ubiquitin-like modifications.
Mol.Cell, 27, 2007
|
|
2JP9
 
 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DGP*DCP*DGP*DC)-3'), DNA (5'-D(P*DGP*DCP*DGP*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-04-30 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
2JPA
 
 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(P*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DG)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-05-01 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
