7ENL
 
 | |
5ENL
 
 | |
6ENL
 
 | |
1NEL
 
 | | FLUORIDE INHIBITION OF YEAST ENOLASE: CRYSTAL STRUCTURE OF THE ENOLASE-MG2+-F--PI COMPLEX AT 2.6-ANGSTROMS RESOLUTION | | Descriptor: | ENOLASE, FLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Lebioda, L, Zhang, E, Lewinski, K, Brewer, M.J. | | Deposit date: | 1993-08-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fluoride inhibition of yeast enolase: crystal structure of the enolase-Mg(2+)-F(-)-Pi complex at 2.6 A resolution.
Proteins, 16, 1993
|
|
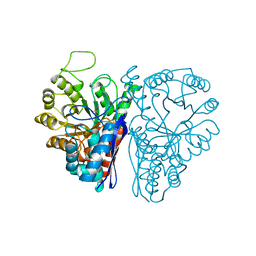
3ENL
 
 | |
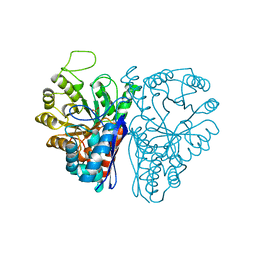
4ENL
 
 | |
4OOD
 
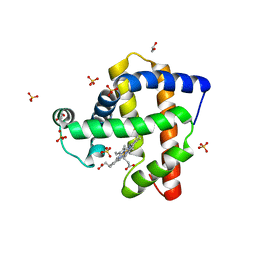 | | Structure of K42Y mutant of sperm whale myoglobin | | Descriptor: | 1,2-ETHANEDIOL, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lebioda, L, Wang, C, Lovelace, L.L. | | Deposit date: | 2014-01-31 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structures of K42N and K42Y sperm whale myoglobins point to an inhibitory role of distal water in peroxidase activity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OF9
 
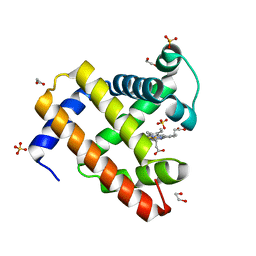 | | Structure of K42N variant of sperm whale myoglobin | | Descriptor: | 1,2-ETHANEDIOL, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lebioda, L, Wang, C, Lovelace, L.L. | | Deposit date: | 2014-01-14 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.241 Å) | | Cite: | Structures of K42N and K42Y sperm whale myoglobins point to an inhibitory role of distal water in peroxidase activity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2ONE
 
 | |
3SDN
 
 | | Structure of G65I sperm whale myoglobin mutant | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Myoglobin, ... | | Authors: | Lebioda, L, Huang, X. | | Deposit date: | 2011-06-09 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Amphitrite ornata Dehaloperoxidase (DHP): Investigations of Structural Factors That Influence the Mechanism of Halophenol Dehalogenation Using "Peroxidase-like" Myoglobin Mutants and "Myoglobin-like" DHP Mutants.
Biochemistry, 50, 2011
|
|
4H07
 
 | | Complex of G65T Myoglobin with Phenol in its Proximal Cavity | | Descriptor: | 1,2-ETHANEDIOL, Myoglobin, PHENOL, ... | | Authors: | Lebioda, L, Lovelace, L.L, Celeste, L.R, Huang, X, Wang, C, Shengfang, S, Dawson, J.H. | | Deposit date: | 2012-09-07 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Complex of myoglobin with phenol bound in a proximal cavity.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3D6I
 
 | | Structure of the Thioredoxin-like Domain of Yeast Glutaredoxin 3 | | Descriptor: | Monothiol glutaredoxin-3, SULFATE ION | | Authors: | Lebioda, L, Gibson, L.M, Dingra, N.N, Outten, C.E. | | Deposit date: | 2008-05-19 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the thioredoxin-like domain of yeast glutaredoxin 3.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
4H0B
 
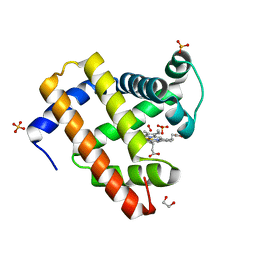 | | Complex of G65T Myoglobin with DMSO in its Distal Cavity | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Myoglobin, ... | | Authors: | Lebioda, L, Lovelace, L.L, Celeste, L.R, Huang, X, Wang, C, Shengfang, S, Dawson, J.H. | | Deposit date: | 2012-09-07 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Complex of myoglobin with phenol bound in a proximal cavity.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1EW6
 
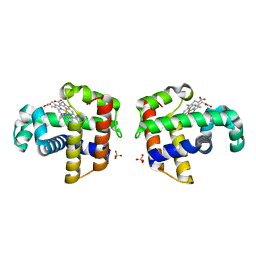 | |
2HPA
 
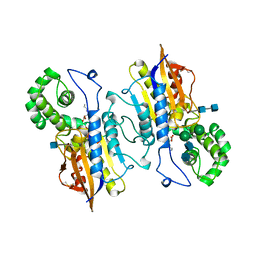 | | STRUCTURAL ORIGINS OF L(+)-TARTRATE INHIBITION OF HUMAN PROSTATIC ACID PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-PROPYL-TARTRAMIC ACID, ... | | Authors: | Lacount, M.W, Handy, G, Lebioda, L. | | Deposit date: | 1998-09-11 | | Release date: | 1998-09-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural origins of L(+)-tartrate inhibition of human prostatic acid phosphatase.
J.Biol.Chem., 273, 1998
|
|
1ELS
 
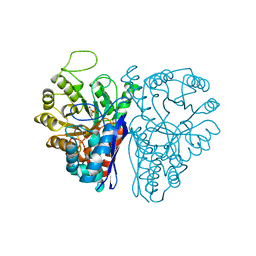 | | CATALYTIC METAL ION BINDING IN ENOLASE: THE CRYSTAL STRUCTURE OF ENOLASE-MN2+-PHOSPHONOACETOHYDROXAMATE COMPLEX AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | ENOLASE, MANGANESE (II) ION, PHOSPHONOACETOHYDROXAMIC ACID | | Authors: | Zhang, E, Hatada, M, Brewer, J.M, Lebioda, L. | | Deposit date: | 1994-04-05 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic metal ion binding in enolase: the crystal structure of an enolase-Mn2+-phosphonoacetohydroxamate complex at 2.4-A resolution.
Biochemistry, 33, 1994
|
|
1TDR
 
 | | EXPRESSION, CHARACTERIZATION, AND CRYSTALLOGRAPHIC ANALYSIS OF TELLUROMETHIONYL DIHYDROFOLATE REDUCTASE | | Descriptor: | CALCIUM ION, CHLORIDE ION, METHOTREXATE, ... | | Authors: | Lewinski, K, Lebioda, L. | | Deposit date: | 1995-04-13 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Expression, characterization and crystallographic analysis of telluromethionyl dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1YPV
 
 | |
1YJ0
 
 | | Crystal Structures of Chicken Annexin V in Complex with Zn2+ | | Descriptor: | Annexin A5, SULFATE ION, ZINC ION | | Authors: | Ortlund, E.A, Chai, G, Genge, B, Wu, L.N.Y, Wuthier, R.E, Lebioda, L. | | Deposit date: | 2005-01-11 | | Release date: | 2005-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structures of Chicken Annexin A5 in Complex with Functional Modifiers Ca2+ and Zn2+ Reveal Zn2+ Induced Formation of Non-Planar Assemblies
Annexins, 1, 2005
|
|
3OB7
 
 | | Human Thymidylate Synthase R163K with Cys 195 covalently modified by Glutathione | | Descriptor: | GLUTATHIONE, PHOSPHATE ION, Thymidylate synthase | | Authors: | Gibson, L.M, Celeste, L.R, Lovelace, L.L, Lebioda, L. | | Deposit date: | 2010-08-06 | | Release date: | 2010-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of human thymidylate synthase R163K with dUMP, FdUMP and glutathione show asymmetric ligand binding.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2EVP
 
 | | The Structures of Thiolate- and Carboxylate-Ligated Ferric H93G Myoglobin: Models for Cytochrome P450 and for Oxyanion-Bound Heme Proteins | | Descriptor: | BETA-MERCAPTOETHANOL, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Qin, J, Perera, R, Lovelace, L.L, Dawson, J.H, Lebioda, L. | | Deposit date: | 2005-10-31 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Thiolate- and Carboxylate-Ligated Ferric H93G Myoglobin: Models for Cytochrome P450 and for Oxyanion-Bound Heme Proteins(,).
Biochemistry, 45, 2006
|
|
2EVK
 
 | | The Structures of Thiolate- and Carboxylate-Ligated Ferric H93G Myoglobin: Models for Cytochrome P450 and for Oxyanion-Bound Heme Proteins | | Descriptor: | ACETIC ACID, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Qin, J, Perera, R, Lovelace, L.L, Dawson, J.H, Lebioda, L. | | Deposit date: | 2005-10-31 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of thiolate- and carboxylate-ligated ferric H93G myoglobin: models for cytochrome P450 and for oxyanion-bound heme proteins.
Biochemistry, 45, 2006
|
|
3OJY
 
 | | Crystal Structure of Human Complement Component C8 | | Descriptor: | CALCIUM ION, Complement component C8 alpha chain, Complement component C8 beta chain, ... | | Authors: | Lovelace, L.L, Cooper, C.L, Sodetz, J.M, Lebioda, L. | | Deposit date: | 2010-08-23 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of human C8 protein provides mechanistic insight into membrane pore formation by complement.
J.Biol.Chem., 286, 2011
|
|
1MXS
 
 | | Crystal structure of 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase from Pseudomonas putida. | | Descriptor: | KDPG Aldolase, SULFATE ION | | Authors: | Watanabe, L, Bell, B.J, Lebioda, L, Rios-Steiner, J.L, Tulinsky, A, Arni, R.K. | | Deposit date: | 2002-10-03 | | Release date: | 2003-09-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase from Pseudomonas putida.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4PGA
 
 | | GLUTAMINASE-ASPARAGINASE FROM PSEUDOMONAS 7A | | Descriptor: | AMMONIUM ION, GLUTAMINASE-ASPARAGINASE, SULFATE ION | | Authors: | Jakob, C.G, Lewinski, K, Lacount, M.W, Roberts, J, Lebioda, L. | | Deposit date: | 1997-01-14 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ion binding induces closed conformation in Pseudomonas 7A glutaminase-asparaginase (PGA): crystal structure of the PGA-SO4(2-)-NH4+ complex at 1.7 A resolution.
Biochemistry, 36, 1997
|
|
