3ZZM
 
 | | Crystal structure of Mycobacterium tuberculosis PurH with a novel bound nucleotide CFAIR, at 2.2 A resolution. | | Descriptor: | 5-(FORMYLAMINO)-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, GLYCEROL, ... | | Authors: | Le Nours, J, Bulloch, E.M.M, Zhang, Z, Greenwood, D.R, Middleditch, M.J, Dickson, J.M.J, Baker, E.N. | | Deposit date: | 2011-09-02 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analyses of a Purine Biosynthetic Enzyme from Mycobacterium Tuberculosis Reveal a Novel Bound Nucleotide.
J.Biol.Chem., 286, 2011
|
|
1HJS
 
 | | Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-1,4-GALACTANASE, ... | | Authors: | Le Nours, J, Ryttersgaard, C, Lo Leggio, L, Ostergaard, P.R, Borchert, T.V, Christensen, L.L.H, Larsen, S. | | Deposit date: | 2003-02-27 | | Release date: | 2003-06-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of Two Fungal Beta-1,4-Galactanases: Searching for the Basis for Temperature and Ph Optimum
Protein Sci., 12, 2003
|
|
1HJU
 
 | | Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-1,4-GALACTANASE, ... | | Authors: | Le Nours, J, Ryttersgaard, C, Lo Leggio, L, Ostergaard, P.R, Borchert, T.V, Christensen, L.L.H, Larsen, S. | | Deposit date: | 2003-02-27 | | Release date: | 2003-06-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Two Fungal Beta-1,4-Galactanases: Searching for the Basis for Temperature and Ph Optimum
Protein Sci., 12, 2003
|
|
1HJQ
 
 | | Structure of two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-1,4-GALACTANASE | | Authors: | Le Nours, J, Ryttersgaard, C, Lo Leggio, L, Ostergaard, P.R, Borchert, T.V, Christensen, L.L.H, Larsen, S. | | Deposit date: | 2003-02-27 | | Release date: | 2003-07-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of Two Fungal Beta-1,4-Galactanases: Searching for the Basis for Temperature and Ph Optimum
Protein Sci., 12, 2003
|
|
2CCR
 
 | | Structure of Beta-1,4-Galactanase | | Descriptor: | CALCIUM ION, TRIETHYLENE GLYCOL, YVFO, ... | | Authors: | Le Nours, J, De Maria, L, Welner, D, Jorgensen, C.T, Christensen, L.L.H, Larsen, S, Lo Leggio, L. | | Deposit date: | 2006-01-18 | | Release date: | 2006-03-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Investigating the Binding of Beta-1,4-Galactan to Bacillus Licheniformis Beta-1,4-Galactanase by Crystallography and Computational Modeling.
Proteins, 75, 2009
|
|
2BVY
 
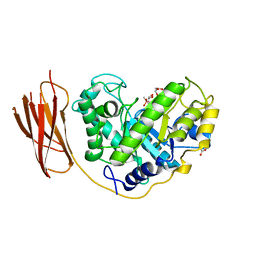 | | The structure and characterization of a modular endo-beta-1,4-mannanase from Cellulomonas fimi | | Descriptor: | ACETATE ION, BETA-1,4-MANNANASE, CACODYLATE ION, ... | | Authors: | Le Nours, J, Anderson, L, Stoll, D, Stalbrand, H, Lo Leggio, L. | | Deposit date: | 2005-07-04 | | Release date: | 2005-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Structure and Characterization of a Modular Endo-Beta-1,4-Mannanase from Cellulomonas Fimi
Biochemistry, 44, 2005
|
|
2BVT
 
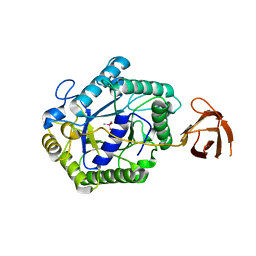 | | The structure of a modular endo-beta-1,4-mannanase from Cellulomonas fimi explains the product specificity of glycoside hydrolase family 26 mannanases. | | Descriptor: | BETA-1,4-MANNANASE, CACODYLATE ION, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Le Nours, J, Anderson, L, Stoll, D, Stalbrand, H, Lo Leggio, L. | | Deposit date: | 2005-07-04 | | Release date: | 2005-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Structure and Characterization of a Modular Endo-Beta-1,4-Mannanase from Cellulomonas Fimi
Biochemistry, 44, 2005
|
|
4BWG
 
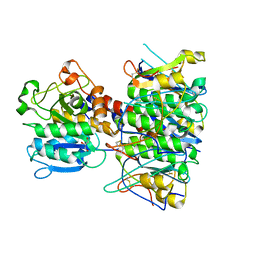 | | Structural basis of subtilase cytotoxin SubAB assembly | | Descriptor: | GLYCEROL, SUBA, SUBTILASE CYTOTOXIN, ... | | Authors: | Le Nours, J, Paton, A.W, Byres, E, Troy, S, Herdman, B.P, Johnson, M.D, Paton, J.C, Rossjohn, J, Beddoe, T. | | Deposit date: | 2013-07-02 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of Subtilase Cytotoxin Subab Assembly.
J.Biol.Chem., 288, 2013
|
|
4A1O
 
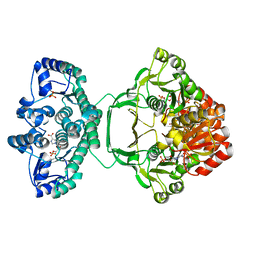 | | Crystal structure of Mycobacterium tuberculosis PurH complexed with AICAR and a novel nucleotide CFAIR, at 2.48 A resolution. | | Descriptor: | 5-(FORMYLAMINO)-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, ... | | Authors: | Le Nours, J, Bulloch, E.M.M, Zhang, Z, Greenwood, D.R, Middleditch, M.J, Dickson, J.M.J, Baker, E.N. | | Deposit date: | 2011-09-17 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural Analyses of a Purine Biosynthetic Enzyme from Mycobacterium Tuberculosis Reveal a Novel Bound Nucleotide.
J.Biol.Chem., 286, 2011
|
|
2J74
 
 | | Structure of Beta-1,4-Galactanase | | Descriptor: | CALCIUM ION, YVFO, beta-D-galactopyranose-(1-4)-beta-D-galactopyranose, ... | | Authors: | Le Nours, J, De Maria, L, Welner, D, Jorgensen, C.T, Christensen, L.L.H, Larsen, S, Lo Leggio, L. | | Deposit date: | 2006-10-06 | | Release date: | 2007-12-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Investigating the Binding of Beta-1,4-Galactan to Bacillus Licheniformis Beta-1,4-Galactanase by Crystallography and Computational Modeling.
Proteins, 75, 2009
|
|
6MWR
 
 | | Recognition of MHC-like molecule | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Le Nours, J, Rossjohn, J. | | Deposit date: | 2018-10-30 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A class of gamma delta T cell receptors recognize the underside of the antigen-presenting molecule MR1.
Science, 366, 2019
|
|
4WW1
 
 | | Crystal structure of human TCR Alpha Chain-TRAV21-TRAJ8 and Beta Chain-TRBV7-8 | | Descriptor: | TCR Alpha Chain-TRAV21-TRAJ8, TCR Beta Chain-TRBV7-8 | | Authors: | Le Nours, J, Praveena, T, Pellicci, D.G, Lim, R.T, Besra, G, Howell, A.R, Godfrey, D.I, Rossjohn, J, Uldrich, A.P. | | Deposit date: | 2014-11-10 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Atypical natural killer T-cell receptor recognition of CD1d-lipid antigens.
Nat Commun, 7, 2016
|
|
4WW2
 
 | | Crystal structure of human TCR Alpha Chain-TRAV21-TRAJ8, Beta Chain-TRBV7-8, Antigen-presenting glycoprotein CD1d, and Beta-2-microglobulin | | Descriptor: | (15Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]tetracos-15-enamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, ... | | Authors: | Le Nours, J, Praveena, T, Pellicci, D, Gherardin, N.A, Lim, R.T, Besra, G, Keshipeddy, S, Richardson, S.K, Howell, A.R, Gras, S, Godfrey, D.I, Rossjohn, J, Uldrich, A.P. | | Deposit date: | 2014-11-10 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Atypical natural killer T-cell receptor recognition of CD1d-lipid antigens.
Nat Commun, 7, 2016
|
|
4WWK
 
 | | Crystal structure of human TCR Alpha Chain-TRAV12-3, Beta Chain-TRBV6-5, Antigen-presenting molecule CD1d, and Beta-2-microglobulin | | Descriptor: | (15Z)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]tetracos-15-enamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, ... | | Authors: | Le Nours, J, Praveena, T, Pellicci, D.G, Gherardin, N.A, Lim, R.T, Besra, G, Keshipeddy, A, Richardson, S.K, Howell, A.R, Gras, S, Godfrey, D.I, Rossjohn, J, Uldrich, A.P. | | Deposit date: | 2014-11-11 | | Release date: | 2016-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Atypical natural killer T-cell receptor recognition of CD1d-lipid antigens.
Nat Commun, 7, 2016
|
|
6BNL
 
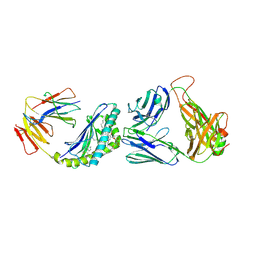 | | Crystal structure of TCR-MHC-like molecule | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Le Nours, J, Rossjohn, J. | | Deposit date: | 2017-11-16 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dual Modifications of alpha-Galactosylceramide Synergize to Promote Activation of Human Invariant Natural Killer T Cells and Stimulate Anti-tumor Immunity.
Cell Chem Biol, 25, 2018
|
|
6BNK
 
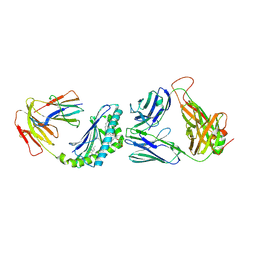 | | Crystal structure of TCR-MHC-like molecule | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Le Nours, J, Rossjohn, J. | | Deposit date: | 2017-11-16 | | Release date: | 2018-04-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Dual Modifications of alpha-Galactosylceramide Synergize to Promote Activation of Human Invariant Natural Killer T Cells and Stimulate Anti-tumor Immunity.
Cell Chem Biol, 25, 2018
|
|
5J1A
 
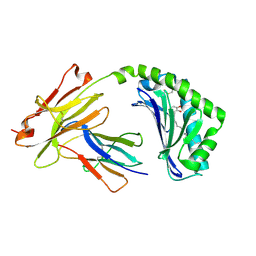 | | Antigen presenting molecule | | Descriptor: | 3-[(8Z,11Z)-pentadeca-8,11-dien-1-yl]benzene-1,2-diol, Beta-2-microglobulin, T-cell surface glycoprotein CD1a | | Authors: | Yongqing, T, Rossjohn, J, Le Nours, J, Marquez, E, Winau, F. | | Deposit date: | 2016-03-29 | | Release date: | 2016-08-03 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | CD1a on Langerhans cells controls inflammatory skin disease.
Nat. Immunol., 17, 2016
|
|
7UFJ
 
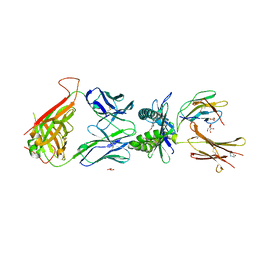 | | Structure of human MR1-ethylvanillin in complex with human MAIT A-F7 TCR | | Descriptor: | 3-ethoxy-4-hydroxybenzaldehyde, ACETATE ION, Beta-2-microglobulin, ... | | Authors: | Wang, C.J, Rossjohn, J, Le Nours, J. | | Deposit date: | 2022-03-22 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Quantitative affinity measurement of small molecule ligand binding to Major Histocompatibility Complex class-I related protein 1 MR1.
J.Biol.Chem., 298, 2022
|
|
7M72
 
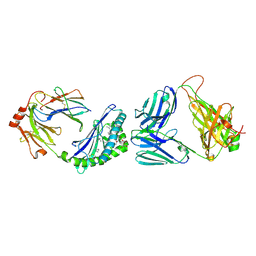 | | MHC-like protein complex structure | | Descriptor: | (3R)-N-[(2S,3R)-1-(alpha-D-galactopyranosyloxy)-3-hydroxy-15-methylhexadecan-2-yl]-3-hydroxyheptadecanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Thirunavukkarasu, P, Le Nours, J, Rossjohn, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Host immunomodulatory lipids created by symbionts from dietary amino acids.
Nature, 600, 2021
|
|
2WZM
 
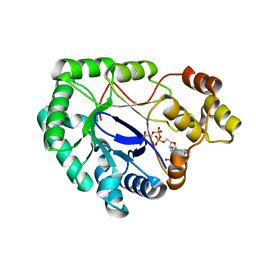 | | Crystal structure of a mycobacterium aldo-keto reductase in its apo and liganded form | | Descriptor: | ALDO-KETO REDUCTASE, [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4S)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Scoble, J, McAlister, A.D, Fulton, Z, Troy, S, Byres, E, Vivian, J.P, Brammananth, R, Wilce, M.C.J, Le Nours, J, Zaker-Tabrizi, L, Coppel, R.L, Crellin, P.K, Rossjohn, J, Beddoe, T. | | Deposit date: | 2009-11-30 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure and Comparative Functional Analyses of a Mycobacterium Aldo-Keto Reductase.
J.Mol.Biol., 398, 2010
|
|
5L2J
 
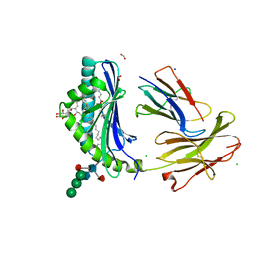 | | Crystal Structure of human CD1b in complex with C36-GMM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-[(2R,3R)-3-hydroxy-2-tetradecyldocosanoyl]-alpha-L-idopyranose, ACETATE ION, ... | | Authors: | Gras, S, Shahine, A, Le Nours, J, Rossjohn, J. | | Deposit date: | 2016-08-02 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | T cell receptor recognition of CD1b presenting a mycobacterial glycolipid.
Nat Commun, 7, 2016
|
|
5L2K
 
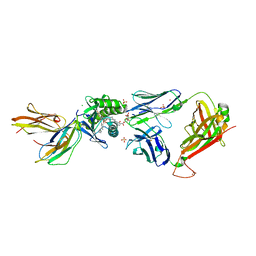 | | Crystal structure of GEM42 TCR-CD1b-GMM complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-[(2R,3R)-3-hydroxy-2-tetradecyldocosanoyl]-alpha-L-idopyranose, Beta-2-microglobulin, ... | | Authors: | Gras, S, Shahine, A, Le Nours, J, Rossjohn, J. | | Deposit date: | 2016-08-02 | | Release date: | 2016-11-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | T cell receptor recognition of CD1b presenting a mycobacterial glycolipid.
Nat Commun, 7, 2016
|
|
2GFT
 
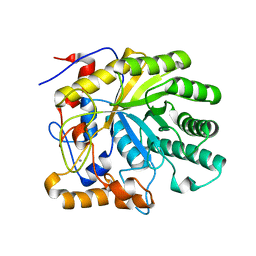 | | Crystal structure of the E263A nucleophile mutant of Bacillus licheniformis endo-beta-1,4-galactanase in complex with galactotriose | | Descriptor: | CALCIUM ION, Glycosyl Hydrolase Family 53, beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Welner, D, Le Nours, J, De Maria, L, Jorgensen, C.T, Christensen, L.L.H, Larsen, S, Lo Leggio, L. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Oligosaccharide binding of Bacillus licheniformis endo-beta-1,4-galactanase
To be Published
|
|
6XNG
 
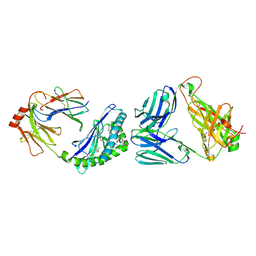 | | MHC-like protein complex structure | | Descriptor: | (3R)-N-[(2S,3R)-1-(alpha-D-galactopyranosyloxy)-3-hydroxyheptadecan-2-yl]-3-hydroxyheptadecanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Thirunavukkarasu, P, Le Nours, J, Rossjohn, J. | | Deposit date: | 2020-07-02 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Host immunomodulatory lipids created by symbionts from dietary amino acids.
Nature, 600, 2021
|
|
8SGM
 
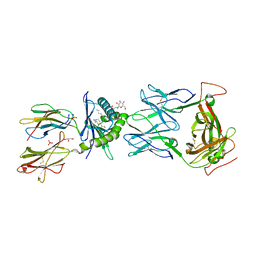 | |
