2UYN
 
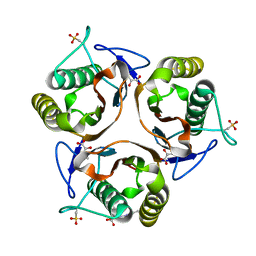 | | Crystal structure of E. coli TdcF with bound 2-ketobutyrate | | Descriptor: | 2-KETOBUTYRIC ACID, PROTEIN TDCF | | Authors: | Burman, J.D, Stevenson, C.E.M, Sawers, R.G, Lawson, D.M. | | Deposit date: | 2007-04-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Escherichia Coli Tdcf, a Member of the Highly Conserved Yjgf/Yer057C/Uk114 Family.
Bmc Struct.Biol., 7, 2007
|
|
2V09
 
 | | SENS161-164DSSN mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Burrell, M.R, Just, V.J, Bowater, L, Fairhurst, S.A, Requena, L, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-05-10 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Oxalate Decarboxylase and Oxalate Oxidase Activities Can be Interchanged with a Specificity Switch of Up to 282 000 by Mutating an Active Site Lid.
Biochemistry, 46, 2007
|
|
2UYJ
 
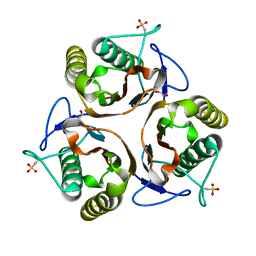 | | Crystal structure of E. coli TdcF with bound ethylene glycol | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN TDCF | | Authors: | Burman, J.D, Stevenson, C.E.M, Sawers, R.G, Lawson, D.M. | | Deposit date: | 2007-04-10 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Crystal Structure of Escherichia Coli Tdcf, a Member of the Highly Conserved Yjgf/Yer057C/Uk114 Family.
Bmc Struct.Biol., 7, 2007
|
|
2UYA
 
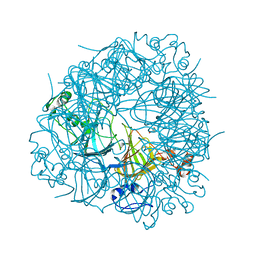 | | DEL162-163 mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
2UYP
 
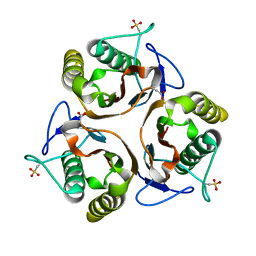 | | Crystal structure of E. coli TdcF with bound propionate | | Descriptor: | PROPANOIC ACID, PROTEIN TDCF | | Authors: | Burman, J.D, Stevenson, C.E.M, Sawers, R.G, Lawson, D.M. | | Deposit date: | 2007-04-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | The Crystal Structure of Escherichia Coli Tdcf, a Member of the Highly Conserved Yjgf/Yer057C/Uk114 Family.
Bmc Struct.Biol., 7, 2007
|
|
2UYB
 
 | | S161A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, MANGANESE (II) ION, ... | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
2UYK
 
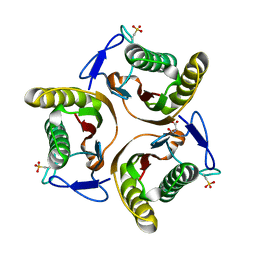 | | Crystal structure of E. coli TdcF with bound serine | | Descriptor: | PROTEIN TDCF, SERINE | | Authors: | Burman, J.D, Stevenson, C.E.M, Sawers, R.G, Lawson, D.M. | | Deposit date: | 2007-04-10 | | Release date: | 2007-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Escherichia Coli Tdcf, a Member of the Highly Conserved Yjgf/Yer057C/Uk114 Family.
Bmc Struct.Biol., 7, 2007
|
|
6F9Q
 
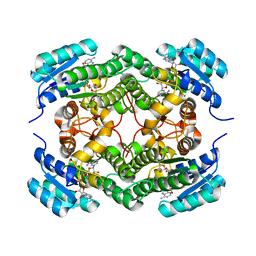 | | Binary complex of a 7S-cis-cis-nepetalactol cyclase from Nepeta mussinii with NAD+ | | Descriptor: | 7S-cis-cis-nepetalactol cyclase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lichman, B.R, Kamileen, M.O, Titchiner, G, Saalbach, G, Stevenson, C.E.M, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2017-12-15 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Uncoupled activation and cyclization in catmint reductive terpenoid biosynthesis.
Nat. Chem. Biol., 15, 2019
|
|
6F8J
 
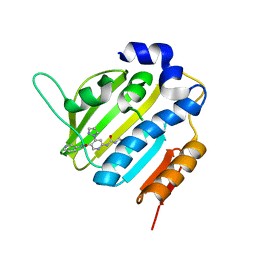 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-(1H-pyrazol-1-yl)-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-4-pyrazol-1-yl-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-13 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F96
 
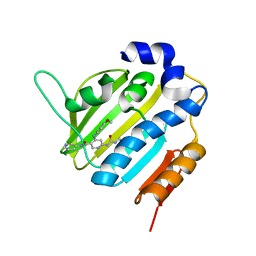 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-[(4-methoxyphenyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-4-[(4-methoxyphenyl)amino]-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F86
 
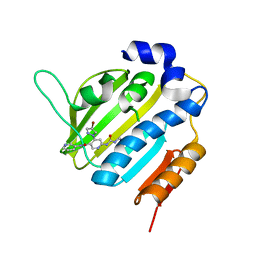 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 4-(4-bromo-1H-pyrazol-1-yl)-6-[(ethylcarbamoyl)amino]-N-(pyridin-3-yl)pyridine-3-carboxamide | | Descriptor: | 4-(4-bromanylpyrazol-1-yl)-6-(ethylcarbamoylamino)-~{N}-pyridin-3-yl-pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-12 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F94
 
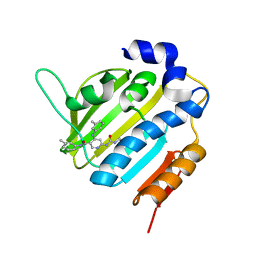 | | Crystal Structure of E. coli GyraseB 24kDa in complex with 6-[(ethylcarbamoyl)amino]-4-[(3-methyphenyl)amino]-N-(3-methyphenyl)pyridine-3-carboxamide | | Descriptor: | 6-(ethylcarbamoylamino)-~{N}-(3-methylphenyl)-4-[(3-methylphenyl)amino]pyridine-3-carboxamide, DNA gyrase subunit B | | Authors: | Narramore, S.K, Stevenson, C.E.M, Lawson, D.M, Maxwell, A, Fishwick, C.W.G. | | Deposit date: | 2017-12-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | New insights into the binding mode of pyridine-3-carboxamide inhibitors of E. coli DNA gyrase.
Bioorg.Med.Chem., 27, 2019
|
|
6F9H
 
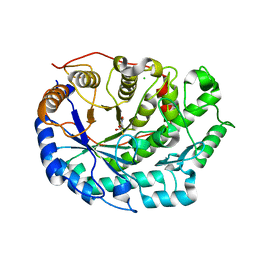 | | Crystal structure of Barley Beta-Amylase complexed with 4-S-alpha-D-glucopyranosyl-(1,4-dideoxy-4-thio-nojirimycin) | | Descriptor: | 1,4-dideoxy-4-thio-nojirimycin, Beta-amylase, CHLORIDE ION, ... | | Authors: | Moncayo, M.A, Rodrigues, L.L, Stevenson, C.E.M, Ruzanski, C, Rejzek, M, Lawson, D.M, Angulo, J, Field, R.A. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis, biological and structural analysis of prospective glycosyl-iminosugar prodrugs: impact on germination
To be published
|
|
6F9L
 
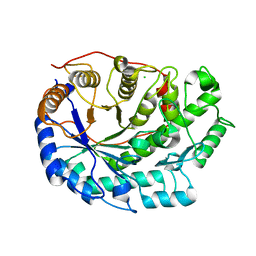 | | Crystal structure of Barley Beta-Amylase complexed with 3-Deoxy-3-fluoro-maltose | | Descriptor: | Beta-amylase, CHLORIDE ION, alpha-D-glucopyranose-(1-4)-3-deoxy-3-fluoro-alpha-D-glucopyranose | | Authors: | Tantanarat, K, Stevenson, C.E.M, Rejzek, M, Lawson, D.M, Field, R.A. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of Barley Beta-Amylase complexed with 3-Deoxy-3-fluoro-maltose
To be published
|
|
6F9J
 
 | | Crystal structure of Barley Beta-Amylase complexed with 4-O-alpha-D-mannopyranosyl-(1-deoxynojirimycin) | | Descriptor: | 1-DEOXYNOJIRIMYCIN, Beta-amylase, CHLORIDE ION, ... | | Authors: | Moncayo, M.A, Rodrigues, L.L, Stevenson, C.E.M, Ruzanski, C, Rejzek, M, Lawson, D.M, Angulo, J, Field, R.A. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Synthesis, biological and structural analysis of prospective glycosyl-iminosugar prodrugs: impact on germination
To be published
|
|
4BQE
 
 | | Arabidopsis thaliana Cytosolic Alpha-1,4-glucan Phosphorylase (PHS2) | | Descriptor: | ALPHA-GLUCAN PHOSPHORYLASE 2,4-GLUCAN PHOSPHORYLASE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | O'Neill, E.C, Rashid, A.M, Stevenson, C.E.M, Hetru, A.C, Gunning, A.P, Rejzek, M, Nepogodiev, S.A, Bornemann, S, Lawson, D.M, Field, R.A. | | Deposit date: | 2013-05-30 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sugar-Coated Sensor Chip and Nanoparticle Surfaces for the in Vitro Enzymatic Synthesis of Starch-Like Materials
Chem.Sci., 5, 2014
|
|
4BWV
 
 | | Structure of Adenosine 5-prime-phosphosulfate Reductase apr-b from Physcomitrella Patens | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHOADENOSINE-PHOSPHOSULPHATE REDUCTASE | | Authors: | Stevenson, C.E.M, Hughes, R.K, McManus, M.T, Lawson, D.M, Kopriva, S. | | Deposit date: | 2013-07-04 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The X-Ray Crystal Structure of Apr-B, an Atypical Adenosine 5-Prime-Phosphosulfate Reductase from Physcomitrella Patens
FEBS Lett., 587, 2013
|
|
4B8X
 
 | | Near atomic resolution crystal structure of Sco5413, a MarR family transcriptional regulator from Streptomyces coelicolor | | Descriptor: | CHLORIDE ION, POSSIBLE MARR-TRANSCRIPTIONAL REGULATOR | | Authors: | Holley, T.A, Stevenson, C.E.M, Bibb, M.J, Lawson, D.M. | | Deposit date: | 2012-08-31 | | Release date: | 2012-10-17 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High Resolution Crystal Structure of Sco5413, a Widespread Actinomycete Marr Family Transcriptional Regulator of Unknown Function.
Proteins, 81, 2013
|
|
3ZQL
 
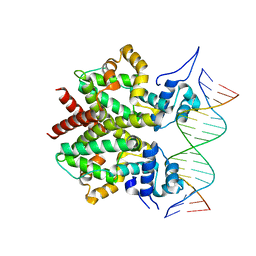 | | DNA-bound form of TetR-like repressor SimR | | Descriptor: | 5'-D(*DTP*TP*CP*GP*TP*AP*CP*GP*CP*CP*GP*TP*AP*DCP *GP*AP*A)-3', 5'-D(*DTP*TP*CP*GP*TP*AP*CP*GP*GP*CP*GP*TP*AP*DCP *GP*AP*A)-3', PUTATIVE REPRESSOR SIMREG2 | | Authors: | Le, T.B.K, Schumacher, M.A, Lawson, D.M, Brennan, R.G, Buttner, M.J. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Crystal Structure of the Tetr Family Transcriptional Repressor Simr Bound to DNA and the Role of a Flexible N-Terminal Extension in Minor Groove Binding.
Nucleic Acids Res., 39, 2011
|
|
4CN6
 
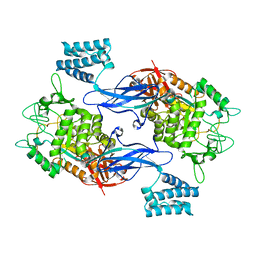 | | GlgE isoform 1 from Streptomyces coelicolor E423A mutant with maltose bound | | Descriptor: | ALPHA-1,4-GLUCAN:MALTOSE-1-PHOSPHATE MALTOSYLTRANSFERASE 1, alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Rashid, A.M, Saalbach, G, Tang, M, Tuukanen, A, Svergun, D.I, Withers, S.G, Lawson, D.M, Bornemann, S. | | Deposit date: | 2014-01-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Insight Into How Streptomyces Coelicolor Maltosyl Transferase Glge Binds Alpha-Maltose 1-Phosphate and Forms a Maltosyl-Enzyme Intermediate.
Biochemistry, 53, 2014
|
|
3ZSS
 
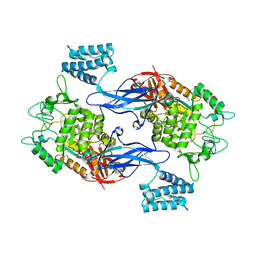 | | Apo form of GlgE isoform 1 from Streptomyces coelicolor | | Descriptor: | PUTATIVE GLUCANOHYDROLASE PEP1A | | Authors: | Syson, K, Stevenson, C.E.M, Rejzek, M, Fairhurst, S.A, Nair, A, Bruton, C.J, Field, R.A, Chater, K.F, Lawson, D.M, Bornemann, S. | | Deposit date: | 2011-06-30 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a Streptomyces Maltosyltransferase Glge: A Homologue of a Genetically Validated Anti-Tuberculosis Target.
J.Biol.Chem., 286, 2011
|
|
4CKK
 
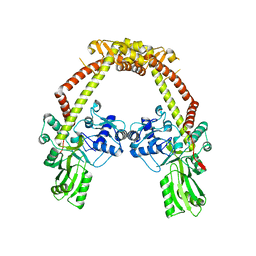 | | Apo structure of 55 kDa N-terminal domain of E. coli DNA gyrase A subunit | | Descriptor: | DNA GYRASE SUBUNIT A | | Authors: | Hearnshaw, S.J, Edwards, M.J, Stevenson, C.E.M, Lawson, D.M, Maxwell, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A New Crystal Structure of the Bifunctional Antibiotic Simocyclinone D8 Bound to DNA Gyrase Gives Fresh Insight Into the Mechanism of Inhibition.
J.Mol.Biol., 426, 2014
|
|
4CKL
 
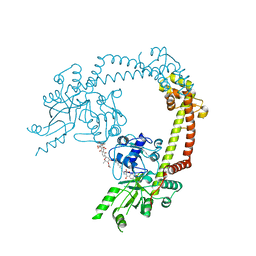 | | Structure of 55 kDa N-terminal domain of E. coli DNA gyrase A subunit with simocyclinone D8 bound | | Descriptor: | DNA GYRASE SUBUNIT A, SIMOCYCLINONE D8 | | Authors: | Hearnshaw, S.J, Edwards, M.J, Stevenson, C.E.M, Lawson, D.M, Maxwell, A. | | Deposit date: | 2014-01-07 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A New Crystal Structure of the Bifunctional Antibiotic Simocyclinone D8 Bound to DNA Gyrase Gives Fresh Insight Into the Mechanism of Inhibition.
J.Mol.Biol., 426, 2014
|
|
3ZPL
 
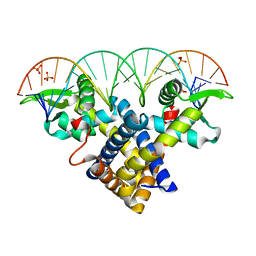 | | Crystal structure of Sco3205, a MarR family transcriptional regulator from Streptomyces coelicolor, in complex with DNA | | Descriptor: | 5'-D(*AP*AP*AP*GP*AP*TP*TP*GP*AP*GP*AP*TP*CP*TP *CP*AP*AP*TP*CP*TP*TP*DT)-3', PHOSPHATE ION, PUTATIVE MARR-FAMILY TRANSCRIPTIONAL REPRESSOR | | Authors: | Stevenson, C.E.M, Assaad, A, Lawson, D.M. | | Deposit date: | 2013-02-28 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Investigation of DNA Sequence Recognition by a Streptomycete Marr Family Transcriptional Regulator Through Surface Plasmon Resonance and X-Ray Crystallography.
Nucleic Acids Res., 41, 2013
|
|
4CN1
 
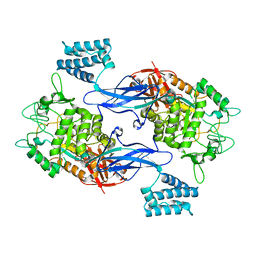 | | GlgE isoform 1 from Streptomyces coelicolor D394A mutant with maltose- 1-phosphate bound | | Descriptor: | ALPHA-1,4-GLUCAN: MALTOSE-1-PHOSPHATE MALTOSYLTRANSFERASE 1, alpha-D-glucopyranose-(1-4)-1-O-phosphono-alpha-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Rashid, A.M, Saalbach, G, Tang, M, Tuukanen, A, Svergun, D.I, Withers, S.G, Lawson, D.M, Bornemann, S. | | Deposit date: | 2014-01-21 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insight Into How Streptomyces Coelicolor Maltosyl Transferase Glge Binds Alpha-Maltose 1-Phosphate and Forms a Maltosyl-Enzyme Intermediate.
Biochemistry, 53, 2014
|
|
