6PXF
 
 | | Structure of human Cathepsin K with an ectosteric inhibitor at 1.85 Angstrom resolution | | Descriptor: | 1,6,6-trimethyl-10,11-dioxo-6,7,8,9,10,11-hexahydrophenanthro[1,2-b]furan-2-sulfonic acid, Cathepsin K | | Authors: | Law, S, Aguda, A.H, Nguyen, N.T, Brayer, G.D, Bromme, D. | | Deposit date: | 2019-07-25 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of human Cathepsin K with an ectosteric inhibitor at 1.85 Angstrom resolution
To Be Published
|
|
5T6U
 
 | | Crystal structure of mouse cathepsin K at 2.9 Angstroms resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cathepsin K, SULFATE ION | | Authors: | Law, S, Aguda, A, Nguyen, N, Brayer, G, Bromme, D. | | Deposit date: | 2016-09-01 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Identification of mouse cathepsin K structural elements that regulate the potency of odanacatib.
Biochem. J., 474, 2017
|
|
5TDI
 
 | | Crystal structure of Cathepsin K with a covalently-linked inhibitor at 1.4 Angstrom resolution. | | Descriptor: | 4-fluoro-N-{1-[(Z)-iminomethyl]cyclopropyl}-N~2~-{(1S)-2,2,2-trifluoro-1-[4'-(methylsulfonyl)[1,1'-biphenyl]-4-yl]ethyl }-L-leucinamide, Cathepsin K | | Authors: | Law, S, Aguda, A, Nguyen, N, Brayer, G, Bromme, D. | | Deposit date: | 2016-09-19 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Identification of mouse cathepsin K structural elements that regulate the potency of odanacatib.
Biochem. J., 474, 2017
|
|
6BKI
 
 | |
6ASH
 
 | | Crystal structure of human Cathepsin K with a non-active site inhibitor at 1.42 Angstrom resolution | | Descriptor: | 2-{[(carbamoylsulfanyl)acetyl]amino}benzoic acid, Cathepsin K | | Authors: | Law, S, Aguda, A, Nguyen, N, Brayer, G, Bromme, D. | | Deposit date: | 2017-08-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.423 Å) | | Cite: | Crystal structure of human Cathepsin K with a non-active site inhibitor at 1.42 Angstrom resolution.
To Be Published
|
|
5IMW
 
 | | Trapped Toxin | | Descriptor: | Intermedilysin | | Authors: | Lawrence, S.L, Feil, S.C, Morton, C.J, Parker, M.W. | | Deposit date: | 2016-03-07 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural Basis for Receptor Recognition by the Human CD59-Responsive Cholesterol-Dependent Cytolysins.
Structure, 24, 2016
|
|
5IMY
 
 | | Trapped Toxin | | Descriptor: | CD59 glycoprotein, Vaginolysin | | Authors: | Lawrence, S.L, Morton, C.J, Parker, M.W. | | Deposit date: | 2016-03-07 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Receptor Recognition by the Human CD59-Responsive Cholesterol-Dependent Cytolysins.
Structure, 24, 2016
|
|
7ZOA
 
 | | cryo-EM structure of CGT ABC transporter in presence of CBG substrate | | Descriptor: | Beta-(1-->2)glucan export ATP-binding/permease protein NdvA, Cyclooctadecakis-(1-2)-(beta-D-glucopyranose) | | Authors: | Jaroslaw, S, Dong, C.N, Frank, L, Na, W, Renato, Z, Seunho, J, Henning, S, Christoph, D. | | Deposit date: | 2022-04-24 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZO8
 
 | | cryo-EM structure of CGT ABC transporter in nanodisc apo state | | Descriptor: | Beta-(1-->2)glucan export ATP-binding/permease protein NdvA, DIUNDECYL PHOSPHATIDYL CHOLINE | | Authors: | Jaroslaw, S, Dong, C.N, Frank, L, Na, W, Renato, Z, Seunho, J, Henning, S, Christoph, D. | | Deposit date: | 2022-04-24 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZO9
 
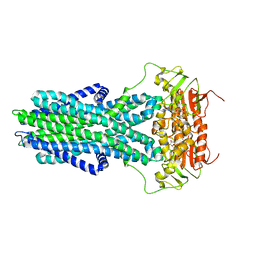 | | cryo-EM structure of CGT ABC transporter in vanadate trapped state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beta-(1-->2)glucan export ATP-binding/permease protein NdvA, VANADATE ION | | Authors: | Jaroslaw, S, Dong, C.N, Frank, L, Na, W, Renato, Z, Seunho, J, Henning, S, Christoph, D. | | Deposit date: | 2022-04-24 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7ZNU
 
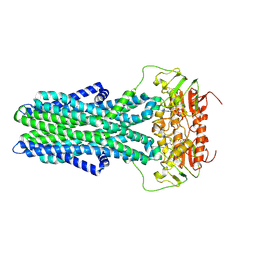 | | cryo-EM structure of CGT ABC transporter in detergent micelle | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beta-(1-->2)glucan export ATP-binding/permease protein NdvA, VANADATE ION | | Authors: | Jaroslaw, S, Dong, C.N, Frank, L, Na, W, Renato, Z, Seunho, J, Henning, S, Christoph, D. | | Deposit date: | 2022-04-22 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Mechanism of cyclic beta-glucan export by ABC transporter Cgt of Brucella.
Nat.Struct.Mol.Biol., 29, 2022
|
|
2AF4
 
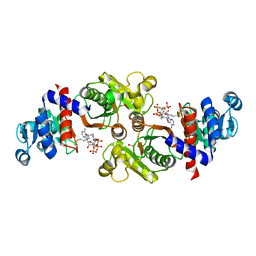 | | Phosphotransacetylase from Methanosarcina thermophila co-crystallized with coenzyme A | | Descriptor: | COENZYME A, Phosphate acetyltransferase | | Authors: | Lawrence, S.H, Luther, K.B, Ferry, J.G, Schindelin, H. | | Deposit date: | 2005-07-25 | | Release date: | 2006-01-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Structural and functional studies suggest a catalytic mechanism for the phosphotransacetylase from Methanosarcina thermophila.
J.Bacteriol., 188, 2006
|
|
2AF3
 
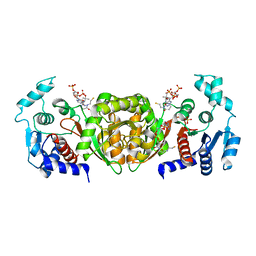 | | Phosphotransacetylase from Methanosarcina thermophila soaked with Coenzyme A | | Descriptor: | COENZYME A, Phosphate acetyltransferase, SULFATE ION | | Authors: | Lawrence, S.H, Luther, K.B, Ferry, J.G, Schindelin, H. | | Deposit date: | 2005-07-25 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional studies suggest a catalytic mechanism for the phosphotransacetylase from Methanosarcina thermophila.
J.Bacteriol., 188, 2006
|
|
1YUK
 
 | | The crystal structure of the PSI/Hybrid domain/ I-EGF1 segment from the human integrin beta2 at 1.8 resolution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, Integrin beta-2 A chain, Integrin beta-2 B chain | | Authors: | Shi, M, Sundramurthy, K, Liu, B, Tan, S.M, Law, S.K, Lescar, J. | | Deposit date: | 2005-02-14 | | Release date: | 2005-07-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of the Plexin-Semaphorin-Integrin Domain/Hybrid Domain/I-EGF1 Segment from the Human Integrin {beta}2 Subunit at 1.8-A Resolution
J.Biol.Chem., 280, 2005
|
|
2P26
 
 | | Structure of the PHE2 and PHE3 fragments of the integrin beta2 subunit | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin beta-2 | | Authors: | Shi, M, Foo, S.Y, Tan, S.M, Mitchell, E.P, Law, S.K.A, Lescar, J. | | Deposit date: | 2007-03-06 | | Release date: | 2007-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A structural hypothesis for the transition between bent and extended conformations of the leukocyte beta2 integrins
J.Biol.Chem., 282, 2007
|
|
2P28
 
 | | Structure of the PHE2 and PHE3 fragments of the integrin beta2 subunit | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin beta-2 | | Authors: | Shi, M, Foo, S.Y, Tan, S.M, Mitchell, E.P, Law, S.K.A, Lescar, J. | | Deposit date: | 2007-03-07 | | Release date: | 2007-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structural hypothesis for the transition between bent and extended conformations of the leukocyte beta2 integrins
J.Biol.Chem., 282, 2007
|
|
5TUN
 
 | | Crystal structure of uninhibited human Cathepsin K at 1.62 Angstrom resolution | | Descriptor: | Cathepsin K | | Authors: | Aguda, A.H, Kruglyak, N, Nguyen, N.T, Law, S, Bromme, D, Brayer, G.D. | | Deposit date: | 2016-11-06 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification of mouse cathepsin K structural elements that regulate the potency of odanacatib.
Biochem. J., 474, 2017
|
|
5DIM
 
 | |
5DHL
 
 | |
5IMT
 
 | | Toxin receptor complex | | Descriptor: | CD59 glycoprotein, COPPER (II) ION, Intermedilysin, ... | | Authors: | Morton, C.J, Lawrence, S.L, Feil, S.C, Parker, M.W. | | Deposit date: | 2016-03-06 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7001 Å) | | Cite: | Structural Basis for Receptor Recognition by the Human CD59-Responsive Cholesterol-Dependent Cytolysins.
Structure, 24, 2016
|
|
6NAL
 
 | |
6XD4
 
 | | CDC-like protein | | Descriptor: | ACETATE ION, Hemolysin, SODIUM ION | | Authors: | Morton, C.J, Parker, M.W, Lawrence, S.L, Johnstone, B.A, Tweten, R.K. | | Deposit date: | 2020-06-09 | | Release date: | 2021-04-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Key Motif in the Cholesterol-Dependent Cytolysins Reveals a Large Family of Related Proteins.
Mbio, 11, 2020
|
|
1QZT
 
 | | Phosphotransacetylase from Methanosarcina thermophila | | Descriptor: | Phosphate acetyltransferase, SULFATE ION | | Authors: | Iyer, P.P, Lawrence, S.H, Luther, K.B, Rajashankar, K.R, Yennawar, H.P, Ferry, J.G, Schindelin, H. | | Deposit date: | 2003-09-17 | | Release date: | 2004-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of phosphotransacetylase from the methanogenic archaeon Methanosarcina thermophila.
STRUCTURE, 12, 2004
|
|
2UZR
 
 | | A transforming mutation in the pleckstrin homology domain of AKT1 in cancer (AKT1-PH_E17K) | | Descriptor: | RAC-alpha serine/threonine-protein kinase | | Authors: | Carpten, J.D, Faber, A.L, Horn, C, Donoho, G.P, Briggs, S.L, Robbins, C.M, Hostetter, G, Boguslawski, S, Moses, T.Y, Savage, S, Uhlik, M, Lin, A, Du, J, Qian, Y.W, Zeckner, D.J, Tucker-Kellogg, G, Touchman, J, Patel, K, Mousses, S, Bittner, M, Schevitz, R, Lai, M.H, Blanchard, K.L, Thomas, J.E. | | Deposit date: | 2007-05-01 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A transforming mutation in the pleckstrin homology domain of AKT1 in cancer.
Nature, 448, 2007
|
|
2UZS
 
 | | A transforming mutation in the pleckstrin homology domain of AKT1 in cancer (AKT1-PH_E17K) | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, RAC-alpha serine/threonine-protein kinase | | Authors: | Carpten, J.D, Faber, A.L, Horn, C, Donoho, G.P, Briggs, S.L, Robbins, C.M, Hostetter, G, Boguslawski, S, Moses, T.Y, Savage, S, Uhlik, M, Lin, A, Du, J, Qian, Y.W, Zeckner, D.J, Tucker-Kellogg, G, Touchman, J, Patel, K, Mousses, S, Bittner, M, Schevitz, R, Lai, M.H, Blanchard, K.L, Thomas, J.E. | | Deposit date: | 2007-05-01 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | A transforming mutation in the pleckstrin homology domain of AKT1 in cancer.
Nature, 448, 2007
|
|
