4PIC
 
 | |
1D2F
 
 | |
1O8R
 
 | |
1ELQ
 
 | | CRYSTAL STRUCTURE OF THE CYSTINE C-S LYASE C-DES | | Descriptor: | L-CYSTEINE/L-CYSTINE C-S LYASE, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Clausen, T, Kaiser, J.T, Steegborn, C, Huber, R, Kessler, D. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-19 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the cystine C-S lyase from Synechocystis: stabilization of cysteine persulfide for FeS cluster biosynthesis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1ELU
 
 | | COMPLEX BETWEEN THE CYSTINE C-S LYASE C-DES AND ITS REACTION PRODUCT CYSTEINE PERSULFIDE. | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-AMINO]-PROPIONIC ACID, L-CYSTEINE/L-CYSTINE C-S LYASE, POTASSIUM ION, ... | | Authors: | Clausen, T, Kaiser, J.T, Steegborn, C, Huber, R, Kessler, D. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the cystine C-S lyase from Synechocystis: stabilization of cysteine persulfide for FeS cluster biosynthesis.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1CL2
 
 | |
1CL1
 
 | |
1CS1
 
 | |
5GL6
 
 | | Msmeg rimP | | Descriptor: | Ribosome maturation factor RimP | | Authors: | Chu, T, Lau, T.C.K. | | Deposit date: | 2016-07-08 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The ribosomal maturation factor P fromMycobacterium smegmatisfacilitates the ribosomal biogenesis by binding to the small ribosomal protein S12.
J. Biol. Chem., 294, 2019
|
|
6ZK7
 
 | | Crystal Structure of human PYROXD1/FAD complex | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pyridine nucleotide-disulfide oxidoreductase domain-containing protein 1 | | Authors: | Meinhart, A, Asanovic, I, Martinez, J, Clausen, T. | | Deposit date: | 2020-06-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The oxidoreductase PYROXD1 uses NAD(P) + as an antioxidant to sustain tRNA ligase activity in pre-tRNA splicing and unfolded protein response.
Mol.Cell, 81, 2021
|
|
1HO4
 
 | | CRYSTAL STRUCTURE OF PYRIDOXINE 5'-PHOSPHATE SYNTHASE IN COMPLEX WITH PYRIDOXINE 5'-PHOSPHATE AND INORGANIC PHOSPHATE | | Descriptor: | PHOSPHATE ION, PYRIDOXINE 5'-PHOSPHATE SYNTHASE, PYRIDOXINE-5'-PHOSPHATE | | Authors: | Garrido-Franco, M, Laber, B, Huber, R, Clausen, T. | | Deposit date: | 2000-12-08 | | Release date: | 2001-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the function of pyridoxine 5'-phosphate synthase.
Structure, 9, 2001
|
|
1HO1
 
 | |
6OWG
 
 | | Structure of a synthetic beta-carboxysome shell, T=4 | | Descriptor: | Ethanolamine utilization protein EutN/carboxysome structural protein Ccml, Microcompartments protein | | Authors: | Sutter, M, Laughlin, T.G, Davies, K.M, Kerfeld, C.A. | | Deposit date: | 2019-05-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of a Syntheticbeta-Carboxysome Shell.
Plant Physiol., 181, 2019
|
|
4TRQ
 
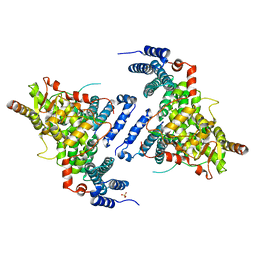 | | Crystal structure of Sac3/Thp1/Sem1 | | Descriptor: | 26S proteasome complex subunit SEM1, Nuclear mRNA export protein SAC3, Nuclear mRNA export protein THP1, ... | | Authors: | Hellerschmied, D, Schneider, S, Kohler, A, Clausen, T. | | Deposit date: | 2014-06-17 | | Release date: | 2015-08-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Nuclear Pore-Associated TREX-2 Complex Employs Mediator to Regulate Gene Expression.
Cell, 162, 2015
|
|
7BII
 
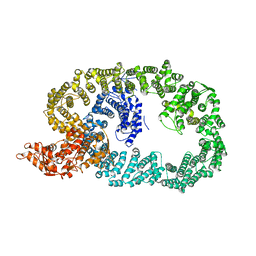 | | Crystal structure of Nematocida HUWE1 | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Grabarczyk, D.B, Petrova, O.A, Meinhart, A, Kessler, D, Clausen, T. | | Deposit date: | 2021-01-12 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.037 Å) | | Cite: | HUWE1 employs a giant substrate-binding ring to feed and regulate its HECT E3 domain.
Nat.Chem.Biol., 17, 2021
|
|
3NWU
 
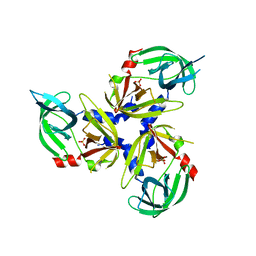 | | Substrate induced remodeling of the active site regulates HtrA1 activity | | Descriptor: | SULFATE ION, Serine protease HTRA1 | | Authors: | Truebestein, L, Tennstaedt, A, Hauske, P, Krojer, T, Kaiser, M, Clausen, T, Ehrmann, M. | | Deposit date: | 2010-07-11 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Substrate-induced remodeling of the active site regulates human HTRA1 activity.
Nat.Struct.Mol.Biol., 18, 2011
|
|
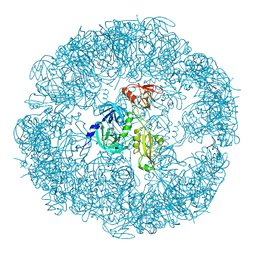
3MH4
 
 | |
3MH5
 
 | | HtrA proteases are activated by a conserved mechanism that can be triggered by distinct molecular cues | | Descriptor: | DIISOPROPYL PHOSPHONATE, Protease do | | Authors: | Krojer, T, Sawa, J, Huber, R, Clausen, T. | | Deposit date: | 2010-04-07 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | HtrA proteases have a conserved activation mechanism that can be triggered by distinct molecular cues
Nat.Struct.Mol.Biol., 17, 2010
|
|
6OWF
 
 | | Structure of a synthetic beta-carboxysome shell, T=3 | | Descriptor: | Ethanolamine utilization protein EutN/carboxysome structural protein Ccml, Microcompartments protein | | Authors: | Sutter, M, Laughlin, T.G, Davies, K.M, Kerfeld, C.A. | | Deposit date: | 2019-05-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a Syntheticbeta-Carboxysome Shell.
Plant Physiol., 181, 2019
|
|
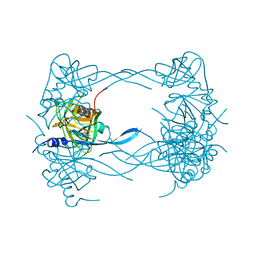
3MH7
 
 | | HtrA proteases are activated by a conserved mechanism that can be triggered by distinct molecular cues | | Descriptor: | 5-mer peptide, Protease do | | Authors: | Krojer, T, Sawa, J, Huber, R, Clausen, T. | | Deposit date: | 2010-04-07 | | Release date: | 2010-06-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.961 Å) | | Cite: | HtrA proteases have a conserved activation mechanism that can be triggered by distinct molecular cues
Nat.Struct.Mol.Biol., 17, 2010
|
|
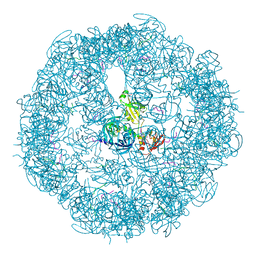
3MH6
 
 | | HtrA proteases are activated by a conserved mechanism that can be triggered by distinct molecular cues | | Descriptor: | DIISOPROPYL PHOSPHONATE, Protease do | | Authors: | Krojer, T, Sawa, J, Huber, R, Clausen, T. | | Deposit date: | 2010-04-07 | | Release date: | 2010-06-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | HtrA proteases have a conserved activation mechanism that can be triggered by distinct molecular cues
Nat.Struct.Mol.Biol., 17, 2010
|
|
3FXU
 
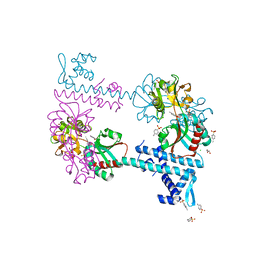 | | Crystal structure of TsaR in complex with its effector p-toluenesulfonate | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Monferrer, D, Tralau, T, Kertesz, M.A, Kikhney, A, Svergun, D, Uson, I. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold
Mol.Microbiol., 75, 2010
|
|
3FZJ
 
 | | TsaR low resolution crystal structure, tetragonal form | | Descriptor: | LysR type regulator of tsaMBCD | | Authors: | Monferrer, D, Tralau, T, Kertesz, M.A, Dix, I, Kikhney, A.G, Svergun, D.I, Uson, I. | | Deposit date: | 2009-01-26 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (7.1 Å) | | Cite: | Structural studies on the full-length LysR-type regulator TsaR from Comamonas testosteroni T-2 reveal a novel open conformation of the tetrameric LTTR fold
Mol.Microbiol., 75, 2010
|
|
7OIK
 
 | |
7OIM
 
 | | Mouse RNF213, with mixed nucleotides bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, E3 ubiquitin-protein ligase RNF213, ... | | Authors: | Grabarczyk, D, Ahel, J, Clausen, T. | | Deposit date: | 2021-05-11 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | E3 ubiquitin ligase RNF213 employs a non-canonical zinc finger active site and is allosterically regulated by ATP
To Be Published
|
|
