2LIR
 
 | | NMR Solution Structure of Yeast Iso-1-cytochrome c Mutant P71H in oxidized states | | Descriptor: | Cytochrome c iso-1, HEME C | | Authors: | Lan, W, Wang, Z, Yang, Z, Zhu, J, Ying, T, Jiang, X, Zhang, X, Wu, H, Liu, M, Tan, X, Cao, C, Huang, Z.X. | | Deposit date: | 2011-08-31 | | Release date: | 2011-12-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational toggling of yeast iso-1-cytochrome C in the oxidized and reduced States.
Plos One, 6, 2011
|
|
2LIT
 
 | | NMR Solution Structure of Yeast Iso-1-cytochrome c Mutant P71H in reduced states | | Descriptor: | Cytochrome c iso-1, HEME C | | Authors: | Lan, W, Wang, Z, Yang, Z, Zhu, J, Ying, T, Jiang, X, Zhang, X, Wu, H, Liu, M, Tan, X, Cao, C, Huang, Z.X. | | Deposit date: | 2011-08-31 | | Release date: | 2011-12-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Conformational toggling of yeast iso-1-cytochrome C in the oxidized and reduced States.
Plos One, 6, 2011
|
|
1G1L
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TDP-GLUCOSE COMPLEX. | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, CITRIC ACID, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naimsmith, J.H. | | Deposit date: | 2000-10-12 | | Release date: | 2000-12-27 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1G23
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). GLUCOSE-1-PHOSPHATE COMPLEX. | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-16 | | Release date: | 2000-12-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1FXO
 
 | |
1FZW
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). APO ENZYME. | | Descriptor: | GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-04 | | Release date: | 2000-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1G2V
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TTP COMPLEX. | | Descriptor: | GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-21 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
1G3L
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TDP-L-RHAMNOSE COMPLEX. | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, SULFATE ION | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naismith, J.H. | | Deposit date: | 2000-10-24 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
5J3F
 
 | | NMR solution structure of [Rp, Rp]-PT dsDNA | | Descriptor: | DNA (5'-D(*CP*GP*(RSG)P*CP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*GP*CP*GP*(RSG)P*CP*CP*G)-3') | | Authors: | Lan, W, Hu, Z, Cao, C. | | Deposit date: | 2016-03-30 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigation into physiological DNA phosphorothioate modification
Sci Rep, 6, 2016
|
|
5J3G
 
 | |
5J3I
 
 | | NMR solution structure of [Sp, Sp]-PT dsDNA | | Descriptor: | DNA (5'-D(*CP*GP*(SSG)P*CP*CP*GP*CP*CP*GP*A)-3'), DNA (5'-D(*TP*CP*GP*GP*CP*GP*(SSG)P*CP*CP*G)-3') | | Authors: | Lan, W, Hu, Z, Cao, C. | | Deposit date: | 2016-03-30 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural investigation into physiological DNA phosphorothioate modification
Sci Rep, 6, 2016
|
|
6YSX
 
 | | Thrombin in complex with 4-amino-N-(5-methylisoxazol-3-yl)benzenesulfonamide (j80) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Hirudin variant-2, ... | | Authors: | Scanlan, W, Heine, A, Klebe, G, Abazi, N. | | Deposit date: | 2020-04-23 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Thrombin in complex with 4-amino-N-(5-methylisoxazol-3-yl)benzenesulfonamide (j80)
To be published
|
|
6YSJ
 
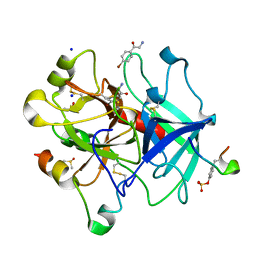 | | Thrombin in complex with 2-amino-1-(4-bromophenyl)ethan-1-one (j10) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-1-(4-bromophenyl)ethanone, DIMETHYL SULFOXIDE, ... | | Authors: | Scanlan, W, Heine, A, Klebe, G, Abazi, N, Paulus, A. | | Deposit date: | 2020-04-22 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Thrombin in complex with 2-amino-1-(4-bromophenyl)ethan-1-one (j10)
To be published
|
|
1U1V
 
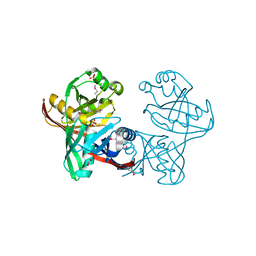 | | Structure and function of phenazine-biosynthesis protein PhzF from Pseudomonas fluorescens 2-79 | | Descriptor: | GLYCEROL, Phenazine biosynthesis protein phzF, SULFATE ION | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6M9L
 
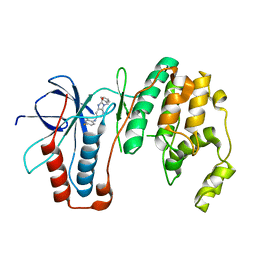 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridine-2-one based p38 MAP Kinase Inhibitors by scaffold hopping - compound 10 | | Descriptor: | 3-benzyl-6-[(2,4-difluorophenyl)amino]-1,3-dihydro-2H-imidazo[4,5-b]pyridin-2-one, Mitogen-activated protein kinase 14 | | Authors: | Lane, W, Okada, K. | | Deposit date: | 2018-08-23 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridin-2-one-Based p38 MAP Kinase Inhibitors: Part 1.
Chemmedchem, 14, 2019
|
|
6E0R
 
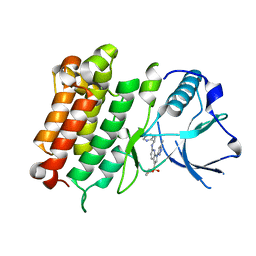 | | hALK in complex with compound 7 N-((1S)-1-(5-fluoropyridin-2-yl)ethyl)-1-(5-methyl-1H-pyrazol-3-yl)-3-(oxetan-3-ylsulfonyl)-1H-pyrrolo[2,3-b]pyridin-6-amine | | Descriptor: | ALK tyrosine kinase receptor, N-[(1S)-1-(5-fluoropyridin-2-yl)ethyl]-1-(5-methyl-1H-pyrazol-3-yl)-3-[(oxetan-3-yl)sulfonyl]-1H-pyrrolo[2,3-b]pyridin-6-amine | | Authors: | Lane, W, Saikatendu, K. | | Deposit date: | 2018-07-06 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Discovery of Potent, Selective, and Brain-Penetrant 1 H-Pyrazol-5-yl-1 H-pyrrolo[2,3- b]pyridines as Anaplastic Lymphoma Kinase (ALK) Inhibitors.
J.Med.Chem., 62, 2019
|
|
6EDL
 
 | | hALK in complex with compound 1 (S)-N-(1-(2,4-difluorophenyl)ethyl)-3-(3-methyl-1H-pyrazol-5-yl)imidazo[1,2-b]pyridazin-6-amine | | Descriptor: | ALK tyrosine kinase receptor, N-[(1S)-1-(2,4-difluorophenyl)ethyl]-3-(5-methyl-1H-pyrazol-3-yl)imidazo[1,2-b]pyridazin-6-amine | | Authors: | Lane, W, Saikatendu, K. | | Deposit date: | 2018-08-09 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Discovery of Potent, Selective, and Brain-Penetrant 1 H-Pyrazol-5-yl-1 H-pyrrolo[2,3- b]pyridines as Anaplastic Lymphoma Kinase (ALK) Inhibitors.
J.Med.Chem., 62, 2019
|
|
7N3L
 
 | | Co-complex CYP46A1 with 0420 (compound 6) | | Descriptor: | 1,2-ETHANEDIOL, Cholesterol 24-hydroxylase, GLYCEROL, ... | | Authors: | Lane, W, Yano, J. | | Deposit date: | 2021-06-01 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Discovery of Novel 3-Piperidinyl Pyridine Derivatives as Highly Potent and Selective Cholesterol 24-Hydroxylase (CH24H) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7N3M
 
 | | Co-complex CYP46A1 with 0431 (compound 17) | | Descriptor: | Cholesterol 24-hydroxylase, N,N-dimethyl-1-[4-(4-methyl-1H-pyrazol-1-yl)pyridin-3-yl]piperidine-4-carboxamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lane, W, Yano, J. | | Deposit date: | 2021-06-01 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Discovery of Novel 3-Piperidinyl Pyridine Derivatives as Highly Potent and Selective Cholesterol 24-Hydroxylase (CH24H) Inhibitors.
J.Med.Chem., 65, 2022
|
|
7N6F
 
 | | Co-complex CYP46A1 with compound 3f | | Descriptor: | (3-fluoroazetidin-1-yl){1-[4-(4-fluorophenyl)pyrimidin-5-yl]piperidin-4-yl}methanone, 1,2-ETHANEDIOL, Cholesterol 24-hydroxylase, ... | | Authors: | Lane, W, Gay, S.C. | | Deposit date: | 2021-06-08 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design and synthesis of aryl-piperidine derivatives as potent and selective PET tracers for cholesterol 24-hydroxylase (CH24H)
Eur.J.Med.Chem., 240, 2022
|
|
6YN3
 
 | | Thrombin in complex with 4-hydroxybenzamide (j89) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-HYDROXYBENZAMIDE, DIMETHYL SULFOXIDE, ... | | Authors: | Scanlan, W, Heine, A, Klebe, G, Abazi, N. | | Deposit date: | 2020-04-10 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Thrombin in complex with 4-hydroxybenzamide (j89)
To be published
|
|
6YMP
 
 | | Thrombin in complex with 3-((5-(tert-butyl)isoxazol-3-yl)methyl)oxetan-3-amine (j54) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(5-tert-butyl-1,2-oxazol-3-yl)methyl]oxetan-3-amine, DIMETHYL SULFOXIDE, ... | | Authors: | Scanlan, W, Heine, A, Klebe, G, Abazi, N. | | Deposit date: | 2020-04-09 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Thrombin in complex with 3-((5-(tert-butyl)isoxazol-3-yl)methyl)oxetan-3-amine (j54)
To be published
|
|
6YQV
 
 | | Thrombin in complex with 5-chlorothiophene-2-sulfonamide (j94) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-chloranylthiophene-2-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Scanlan, W, Heine, A, Klebe, G, Abazi, N. | | Deposit date: | 2020-04-18 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Thrombin in complex with 5-chlorothiophene-2-sulfonamide (j94)
To be published
|
|
2FXO
 
 | | Structure of the human beta-myosin S2 fragment | | Descriptor: | Myosin heavy chain, cardiac muscle beta isoform | | Authors: | Blankenfeldt, W, Thoma, N.H, Wray, J.S, Gautel, M, Schlichting, I. | | Deposit date: | 2006-02-06 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human cardiac {beta}-myosin II S2-{Delta} provide insight into the functional role of the S2 subfragment
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2FXM
 
 | | Structure of the human beta-myosin S2 fragment | | Descriptor: | MERCURY (II) ION, Myosin heavy chain, cardiac muscle beta isoform | | Authors: | Blankenfeldt, W, Thoma, N.H, Wray, J.S, Gautel, M, Schlichting, I. | | Deposit date: | 2006-02-06 | | Release date: | 2006-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of human cardiac {beta}-myosin II S2-{Delta} provide insight into the functional role of the S2 subfragment
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
