7T4J
 
 | | Crystal Structure of EGFR_D770_N771insNPG/V948R in complex with TAK-788 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Epidermal growth factor receptor, ... | | Authors: | Skene, R.J, Lane, W, Hu, Y. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of mobocertinib, a potent, oral inhibitor of EGFR exon 20 insertion mutations in non-small cell lung cancer.
Bioorg.Med.Chem.Lett., 80, 2022
|
|
7T4I
 
 | | Crystal Structure of wild type EGFR in complex with TAK-788 | | Descriptor: | Epidermal growth factor receptor, propan-2-yl 2-[[4-[2-(dimethylamino)ethyl-methyl-amino]-2-methoxy-5-(propanoylamino)phenyl]amino]-4-(1-methylindol-3-yl)pyrimidine-5-carboxylate | | Authors: | Skene, R.J, Lane, W. | | Deposit date: | 2021-12-10 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery of mobocertinib, a potent, oral inhibitor of EGFR exon 20 insertion mutations in non-small cell lung cancer.
Bioorg.Med.Chem.Lett., 80, 2022
|
|
7OZQ
 
 | | Crystal structure of archaeal L7Ae bound to eukaryotic kink-loop | | Descriptor: | 50S ribosomal protein L7Ae, ACETATE ION, CALCIUM ION, ... | | Authors: | Hoefler, S, Lukat, P, Carlomagno, T, Blankenfeldt, W. | | Deposit date: | 2021-06-28 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Eukaryotic Box C/D methylation machinery has two non-symmetric protein assembly sites.
Sci Rep, 11, 2021
|
|
1NMS
 
 | | Caspase-3 tethered to irreversible inhibitor | | Descriptor: | 5-[4-(1-CARBOXYMETHYL-2-OXO-PROPYLCARBAMOYL)-BENZYLSULFAMOYL]-2-HYDROXY-BENZOIC ACID, Caspase-3 | | Authors: | Erlanson, D.A, Lam, J, Wiesmann, C, Luong, T.N, Simmons, R.L, DeLano, W.L, Choong, I.C, Flanagan, W.M, Lee, D, O'Brian, T. | | Deposit date: | 2003-01-10 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In situ assembly of enzyme inhibitors using extended tethering.
Nat.Biotechnol., 21, 2003
|
|
1NME
 
 | | Structure of Casp-3 with tethered salicylate | | Descriptor: | 2-HYDROXY-5-(2-MERCAPTO-ETHYLSULFAMOYL)-BENZOIC ACID, 3-(2-MERCAPTO-ACETYLAMINO)-4-OXO-PENTANOIC ACID, Caspase-3 | | Authors: | Erlanson, D.A, Lam, J, Wiesmann, C, Luong, T.N, Simmons, B, DeLano, W, Choong, I.C, Flanagan, M, Lee, D, O'Brian, T. | | Deposit date: | 2003-01-09 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In situ assembly of enzyme inhibitors using extended tethering.
Nat.Biotechnol., 21, 2003
|
|
5JGJ
 
 | | Crystal structure of GtmA | | Descriptor: | UbiE/COQ5 family methyltransferase, putative | | Authors: | Dolan, S.K, Bock, T, Hering, V, Jones, G.W, Blankenfeldt, W, Doyle, S. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-01 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural, mechanistic and functional insight into gliotoxinbis-thiomethylation inAspergillus fumigatus.
Open Biol, 7, 2017
|
|
1NMQ
 
 | | Extendend Tethering: In Situ Assembly of Inhibitors | | Descriptor: | 3-(3-{2-[(5-METHANESULFONYL-THIOPHENE-2-CARBONYL)-AMINO]-ETHYLDISULFANYLMETHYL}- BENZENESULFONYLAMINO)-4-OXO-PENTANOIC ACID, Caspase-3 | | Authors: | Erlanson, D.A, Lam, J, Wiesmann, C, Luong, T.N, Simmons, R.L, DeLano, W, Choong, I.C, Flanagan, M, Lee, D, O'Brian, T. | | Deposit date: | 2003-01-10 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | In situ assembly of enzyme inhibitors using extended tethering.
Nat.Biotechnol., 21, 2003
|
|
7P4U
 
 | |
4QGS
 
 | |
7QY3
 
 | | Crystal structure of the halohydrin dehalogenase HheG D114C mutant cross-linked with BMOE | | Descriptor: | 1,1'-ethane-1,2-diylbis(1H-pyrrole-2,5-dione), Putative oxidoreductase, SULFATE ION | | Authors: | Henke, S, Blankenfeldt, W, Schallmey, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Biocatalytically active and stable cross-linked enzyme crystals of halohydrin dehalogenase HheG by protein engineering
Chemcatchem, 2022
|
|
7QA0
 
 | |
7QA3
 
 | |
7QAV
 
 | |
1G20
 
 | | MGATP-BOUND AND NUCLEOTIDE-FREE STRUCTURES OF A NITROGENASE PROTEIN COMPLEX BETWEEN LEU127DEL-FE PROTEIN AND THE MOFE PROTEIN | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Chiu, H.-J, Peters, J.W, Lanzilotta, W.N, Ryle, M.J, Seefeldt, L.C, Howard, J.B, Rees, D.C. | | Deposit date: | 2000-10-16 | | Release date: | 2001-01-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MgATP-Bound and nucleotide-free structures of a nitrogenase protein complex between the Leu 127 Delta-Fe-protein and the MoFe-protein.
Biochemistry, 40, 2001
|
|
7OC6
 
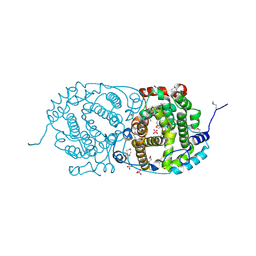 | | Selenomethionine derivative of alpha-humulene synthase AsR6 from Sarocladium schorii | | Descriptor: | Alpha-humulene synthase asR6, GLYCEROL, SULFATE ION, ... | | Authors: | Schotte, C, Lukat, P, Deuschmann, A, Blankenfeldt, W, Cox, R.J. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-07 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Understanding and Engineering the Stereoselectivity of Humulene Synthase.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7OC4
 
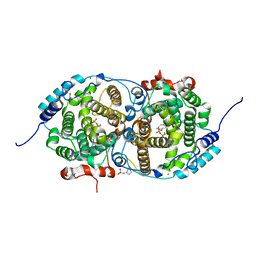 | | Alpha-humulene synthase AsR6 from Sarocladium schorii in complex with thiolodiphosphate and a cyclized reaction product. | | Descriptor: | (1E,4E,8E)-2,6,6,9-Tetramethyl-1,4-8-cycloundecatriene, (R,R)-2,3-BUTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Schotte, C, Lukat, P, Deuschmann, A, Blankenfeldt, W, Cox, R.J. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Understanding and Engineering the Stereoselectivity of Humulene Synthase.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
1G21
 
 | | MGATP-BOUND AND NUCLEOTIDE-FREE STRUCTURES OF A NITROGENASE PROTEIN COMPLEX BETWEEN LEU127DEL-FE PROTEIN AND THE MOFE PROTEIN | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Chiu, H.-J, Peters, J.W, Lanzilotta, W.N, Ryle, M.J, Seefeldt, L.C, Howard, J.B, Rees, D.C. | | Deposit date: | 2000-10-16 | | Release date: | 2001-01-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | MgATP-Bound and nucleotide-free structures of a nitrogenase protein complex between the Leu 127 Delta-Fe-protein and the MoFe-protein.
Biochemistry, 40, 2001
|
|
7OC5
 
 | | Alpha-humulene synthase AsR6 from Sarocladium schorii | | Descriptor: | Alpha-humulene synthase AsR6, MAGNESIUM ION, ZINC ION | | Authors: | Schotte, C, Lukat, P, Deuschmann, A, Blankenfeldt, W, Cox, R.J. | | Deposit date: | 2021-04-26 | | Release date: | 2021-07-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Understanding and Engineering the Stereoselectivity of Humulene Synthase.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7NBW
 
 | |
6XI2
 
 | | Apo form of POMGNT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA, ALA-GLY-ALA-GLY-ALA-ALA-ALA-ALA-ALA-ALA, ... | | Authors: | Halmo, S.M, Yeh, J, Wells, L, Moremen, K.W, Lanzilotta, W.N. | | Deposit date: | 2020-06-19 | | Release date: | 2021-04-21 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystal structures of beta-1,4-N-acetylglucosaminyltransferase 2: structural basis for inherited muscular dystrophies.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6YHL
 
 | |
1FEH
 
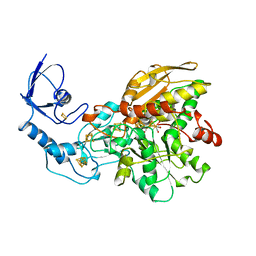 | | FE-ONLY HYDROGENASE FROM CLOSTRIDIUM PASTEURIANUM | | Descriptor: | 2 IRON/2 SULFUR/5 CARBONYL/2 WATER INORGANIC CLUSTER, FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Peters, J.W, Lanzilotta, W.N, Lemon, B.J, Seefeldt, L.C. | | Deposit date: | 1998-10-28 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 angstrom resolution.
Science, 282, 1998
|
|
2O2F
 
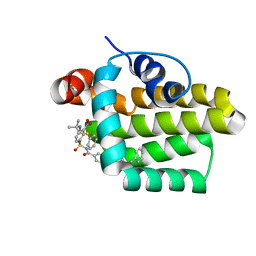 | | Solution structure of the anti-apoptotic protein Bcl-2 in complex with an acyl-sulfonamide-based ligand | | Descriptor: | 4-(4-BENZYL-4-METHOXYPIPERIDIN-1-YL)-N-[(4-{[1,1-DIMETHYL-2-(PHENYLTHIO)ETHYL]AMINO}-3-NITROPHENYL)SULFONYL]BENZAMIDE, Apoptosis regulator Bcl-2 | | Authors: | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | Deposit date: | 2006-11-29 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
1GG6
 
 | | CRYSTAL STRUCTURE OF GAMMA CHYMOTRYPSIN WITH N-ACETYL-PHENYLALANINE TRIFLUOROMETHYL KETONE BOUND AT THE ACTIVE SITE | | Descriptor: | 1,1,1-TRIFLUORO-3-ACETAMIDO-4-PHENYL BUTAN-2-ONE(N-ACETYL-L-PHENYLALANYL TRIFLUOROMETHYL KETONE), 1,2-ETHANEDIOL, GAMMA CHYMOTRYPSIN, ... | | Authors: | Neidhart, D, Wei, Y, Cassidy, C, Lin, J, Cleland, W.W, Frey, P.A. | | Deposit date: | 2000-07-31 | | Release date: | 2000-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Correlation of low-barrier hydrogen bonding and oxyanion binding in transition state analogue complexes of chymotrypsin.
Biochemistry, 40, 2001
|
|
1GGD
 
 | | CRYSTAL STRUCTURE OF GAMMA CHYMOTRYPSIN WITH N-ACETYL-LEUCIL-PHENYLALANINE ALDEHYDE BOUND AT THE ACTIVE SITE | | Descriptor: | 2-ACETYLAMINO-4-METHYL-PENTANOIC ACID (1-FORMYL-2-PHENYL-ETHYL)-AMIDE, GAMMA CHYMOTRYPSIN, SULFATE ION | | Authors: | Neidhart, D, Wei, Y, Cassidy, C, Lin, J, Cleland, W.W, Frey, P.A. | | Deposit date: | 2000-08-14 | | Release date: | 2000-09-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Correlation of low-barrier hydrogen bonding and oxyanion binding in transition state analogue complexes of chymotrypsin.
Biochemistry, 40, 2000
|
|
