3NE7
 
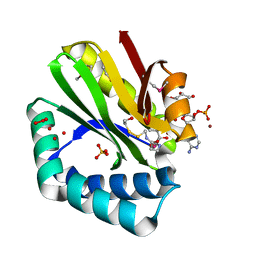 | | Crystal structure of paia n-acetyltransferase from thermoplasma acidophilum in complex with coenzyme a | | Descriptor: | ACETYLTRANSFERASE, BETA-MERCAPTOETHANOL, COENZYME A, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-08 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production.
Proteins, 79, 2011
|
|
3FHR
 
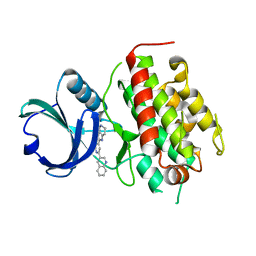 | | High resolution crystal structure of mitogen-activated protein kinase-activated protein kinase 3 (MK3)-inhibitor complex | | Descriptor: | 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MAP kinase-activated protein kinase 3 | | Authors: | Cheng, R.K.Y, Barker, J, Palan, S, Felicetti, B, Whittaker, M, Hesterkamp, T. | | Deposit date: | 2008-12-10 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structure of human Mapkap kinase 3 in complex with a high affinity ligand
Protein Sci., 19, 2010
|
|
4JI1
 
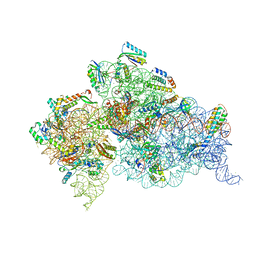 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, RIBOSOMAL PROTEIN S10, ... | | Authors: | Demirci, H, Wang, L, Murphy IV, F, Murphy, E, Carr, J, Blanchard, S, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-03-05 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.144 Å) | | Cite: | The central role of protein S12 in organizing the structure of the decoding site of the ribosome.
Rna, 19, 2013
|
|
2PKF
 
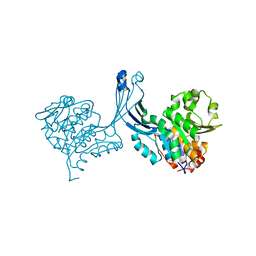 | | Crystal structure of M tuberculosis Adenosine Kinase (apo) | | Descriptor: | Adenosine kinase | | Authors: | Reddy, M.C.M, Palaninathan, S.K, Shetty, N.D, Owen, J.L, Watson, M.D, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-04-17 | | Release date: | 2007-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High resolution crystal structures of Mycobacterium tuberculosis adenosine kinase: insights into the mechanism and specificity of this novel prokaryotic enzyme
J.Biol.Chem., 282, 2007
|
|
1JYA
 
 | | Crystal Structure of SycE | | Descriptor: | YOPE regulator | | Authors: | Ghosh, P, Birtalan, S. | | Deposit date: | 2001-09-11 | | Release date: | 2001-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structure of the Yersinia type III secretory system chaperone SycE.
Nat.Struct.Biol., 8, 2001
|
|
1XTY
 
 | | Crystal structure of Sulfolobus solfataricus peptidyl-tRNA hydrolase | | Descriptor: | Peptidyl-tRNA hydrolase, SULFATE ION | | Authors: | Fromant, M, Schmitt, E, Mechulam, Y, Lazennec, C, Plateau, P, Blanquet, S. | | Deposit date: | 2004-10-25 | | Release date: | 2005-03-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure at 1.8 A resolution and identification of active site residues of Sulfolobus solfataricus peptidyl-tRNA hydrolase.
Biochemistry, 44, 2005
|
|
3N87
 
 | | Crystal structure of 3-dehydroquinate dehydratase from Mycobacterium tuberculosis in complex with inhibitor 3 | | Descriptor: | (1R,4R,5R)-1,4,5-trihydroxy-3-[3-(phenylcarbonyl)phenyl]cyclohex-2-ene-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Dias, M.V.B, Snee, W.C, Bromfield, K.M, Payne, R, Palaninathan, S.K, Ciulli, A, Howard, N.I, Abell, C, Sacchettini, J.C, Blundell, T.L. | | Deposit date: | 2010-05-27 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
4JI5
 
 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, RIBOSOMAL PROTEIN S10, ... | | Authors: | Demirci, H, Wang, L, Murphy IV, F, Murphy, E, Carr, J, Blanchard, S, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-03-05 | | Release date: | 2013-11-06 | | Last modified: | 2013-12-04 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | The central role of protein S12 in organizing the structure of the decoding site of the ribosome.
Rna, 19, 2013
|
|
3GAA
 
 | |
2XHD
 
 | | Crystal structure of N-((2S)-5-(6-fluoro-3-pyridinyl)-2,3-dihydro-1H- inden-2-yl)-2-propanesulfonamide in complex with the ligand binding domain of the human GluA2 receptor | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, N-[(2S)-5-(6-FLUORO-3-PYRIDINYL)-2,3-DIHYDRO-1H-INDEN-2-YL]-2-PROPANESULFONAMIDE, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Andreotti, D, Ballantine, S, Bax, B.D, Harris, A.J, Harker, A.J, Lund, J, Melarange, R, Mingardi, A, Mookherjee, C, Mosley, J, Neve, M, Oliosi, B, Profeta, R, Smith, K.J, Smith, P.W, Spada, S, Thewlis, K.M, Yusaf, S.P. | | Deposit date: | 2010-06-14 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of N-[(2S)-5-(6-Fluoro-3-Pyridinyl)-2,3-Dihydro-1H-Inden-2-Yl]-2-Propanesulfonamide, a Novel Clinical Ampa Receptor Positive Modulator.
J.Med.Chem., 53, 2010
|
|
5KCS
 
 | | Cryo-EM structure of the Escherichia coli 70S ribosome in complex with antibiotic Evernimycin, mRNA, TetM and P-site tRNA at 3.9A resolution | | Descriptor: | (2R,3R,4R,6S)-6-{[(2R,3aR,4R,4'R,5'S,6S,6'R,7S,7aR)-6-{[(2S,3R,4R,5S,6R)-2-{[(2R,3S,4S,5S,6S)-6-({(2R,3aS,3a'R,6S,7R,7' R,7aS,7a'S)-7'-[(2,4-dihydroxy-6-methylbenzoyl)oxy]-7-hydroxyoctahydro-4H-2,4'-spirobi[[1,3]dioxolo[4,5-c]pyran]-6-yl}ox y)-4-hydroxy-5-methoxy-2-(methoxymethyl)tetrahydro-2H-pyran-3-yl]oxy}-3-hydroxy-5-methoxy-6-methyltetrahydro-2H-pyran-4- yl]oxy}-4',7-dihydroxy-4,6',7a-trimethyloctahydro-4H-spiro[1,3-dioxolo[4,5-c]pyran-2,2'-pyran]-5'-yl]oxy}-4-{[(2R,4S,5R, 6S)-5-methoxy-4,6-dimethyl-4-nitrotetrahydro-2H-pyran-2-yl]oxy}-2-methyltetrahydro-2H-pyran-3-yl 3,5-dichloro-4-hydroxy-2-methoxy-6-methylbenzoate (non-preferred name), 16S Ribosomal RNA, ... | | Authors: | Arenz, S, Juette, M.F, Graf, M, Nguyen, F, Huter, P, Polikanov, Y.S, Blanchard, S.C, Wilson, D.N. | | Deposit date: | 2016-06-06 | | Release date: | 2016-08-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of the orthosomycin antibiotics avilamycin and evernimicin in complex with the bacterial 70S ribosome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6WA3
 
 | | Solution NMR structure of the myristoylated feline immunodeficiency virus matrix protein | | Descriptor: | MYRISTIC ACID, Matrix protein | | Authors: | Brown, J.B, Summers, H.R, Brown, L.A, Marchant, J, Canova, P.N, O'Hern, C.T, Abbott, S, Nyaunu, C, Maxwell, S, Johnson, T, Moser, M, Ablan, S.D, Carter, H, Freed, E.O, Summers, M.F. | | Deposit date: | 2020-03-24 | | Release date: | 2020-07-22 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural and Mechanistic Studies of the Rare Myristoylation Signal of the Feline Immunodeficiency Virus.
J.Mol.Biol., 432, 2020
|
|
5KCR
 
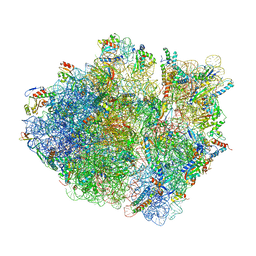 | | Cryo-EM structure of the Escherichia coli 70S ribosome in complex with antibiotic Avilamycin C, mRNA and P-site tRNA at 3.6A resolution | | Descriptor: | (2R,3S,4R,6S)-4-hydroxy-6-{[(2R,3aR,4R,4'R,5'S,6S,6'R,7aR)-4'-hydroxy-6-{[(2S,3R,4R,5S,6R)-3-hydroxy-2-{[(2R,3S,4S,5S,6S)-4-hydroxy-6-({(2R,3aS,3a'R,6S,6'R,7R,7'R,7aR,7a'R)-7'-hydroxy-7'-[(1S)-1-hydroxyethyl]-6'-methyl-7-[(2-methylpropanoyl)oxy]octahydro-4H-2,4'-spirobi[[1,3]dioxolo[4,5-c]pyran]-6-yl}oxy)-5-methoxy-2-(methoxymethyl)tetrahydro-2H-pyran-3-yl]oxy}-5-methoxy-6-methyltetrahydro-2H-pyran-4-yl]oxy}-4,6',7a-trimethyloctahydro-4H-spiro[1,3-dioxolo[4,5-c]pyran-2,2'-pyran]-5'-yl]oxy}-2-methyltetrahydro-2H-pyran-3-yl 3,5-dichloro-4-hydroxy-2-methoxy-6-methylbenzoate (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Arenz, S, Juette, M.F, Graf, M, Nguyen, F, Huter, P, Polikanov, Y.S, Blanchard, S.C, Wilson, D.N. | | Deposit date: | 2016-06-06 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of the orthosomycin antibiotics avilamycin and evernimicin in complex with the bacterial 70S ribosome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4N3P
 
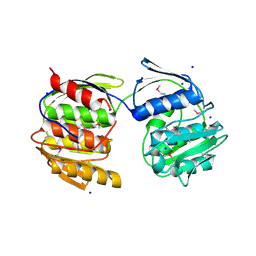 | | Crystal Structure of De Novo designed Serine Hydrolase OSH18, Northeast Structural Genomics Consortium (NESG) Target OR396 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Kuzin, A, Lew, S, Rajagopalan, S, Seetharaman, J, Mao, L, Xiao, R, Kogan, S, Maglaqui, M, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-10-07 | | Release date: | 2013-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal Structure of De Novo designed Serine Hydrolase OSH18, Northeast Structural Genomics Consortium (NESG) Target OR396
To be Published
|
|
4JI7
 
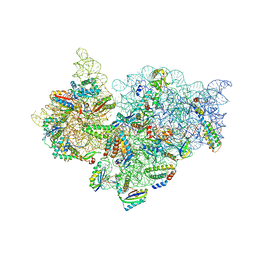 | | Crystal Structure of 30S ribosomal subunit from Thermus thermophilus | | Descriptor: | 16S rRNA, MAGNESIUM ION, RIBOSOMAL PROTEIN S10, ... | | Authors: | Demirci, H, Wang, L, Murphy IV, F, Murphy, E, Carr, J, Blanchard, S, Jogl, G, Dahlberg, A.E, Gregory, S.T. | | Deposit date: | 2013-03-05 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The central role of protein S12 in organizing the structure of the decoding site of the ribosome.
Rna, 19, 2013
|
|
2XXI
 
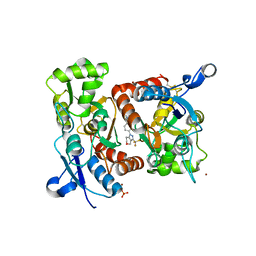 | | Crystal structure of 1-((4-(3-(trifluoromethyl)-6,7-dihydropyrano(4,3- c(pyrazol-1(4H)-yl)phenyl)methyl)-2-pyrrolidinone in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 1.6A resolution. | | Descriptor: | 1-({4-[3-(TRIFLUOROMETHYL)-6,7-DIHYDROPYRANO[4,3-C]PYRAZOL-1(4H)-YL]PHENYL}METHYL)-2-PYRROLIDINONE, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-10 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
2FI9
 
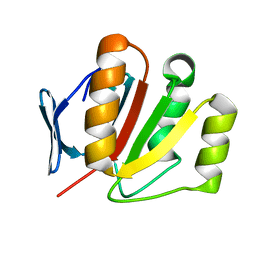 | | The crystal structure of an outer membrane protein from the Bartonella henselae | | Descriptor: | Outer membrane protein | | Authors: | Zhang, R, Hatzos, C, Clancy, S, Collart, F, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-12-28 | | Release date: | 2006-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8A crystal structure of an outer membrane protein from the Bartonella henselae
To be Published
|
|
2XXH
 
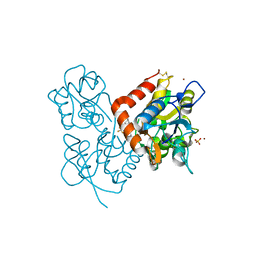 | | Crystal structure of 1-(4-(2-oxo-2-(1-pyrrolidinyl)ethyl)phenyl)-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazole in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 1.5A resolution. | | Descriptor: | 1-{4-[2-OXO-2-(1-PYRROLIDINYL)ETHYL]PHENYL}-3-( TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOLE, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-10 | | Release date: | 2011-04-06 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
2XX7
 
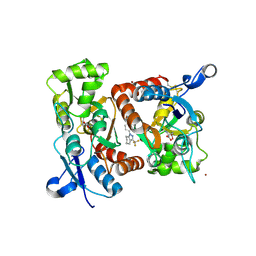 | | Crystal structure of 1-(4-(1-pyrrolidinylcarbonyl)phenyl)-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazole in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 2.2A resolution. | | Descriptor: | 1-[4-(1-PYRROLIDINYLCARBONYL)PHENYL]-3-(TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOLE, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-09 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
2XX8
 
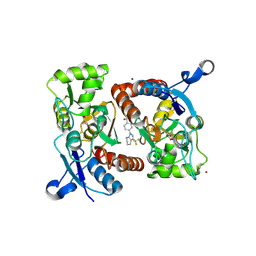 | | Crystal structure of N,N-dimethyl-4-(3-(trifluoromethyl)-4,5,6,7- tetrahydro-1H-indazol-1-yl)benzamide in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 2.2A resolution. | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, N,N-DIMETHYL-4-[3-(TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOL-1-YL]BENZAMIDE, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-09 | | Release date: | 2011-04-27 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
2PKN
 
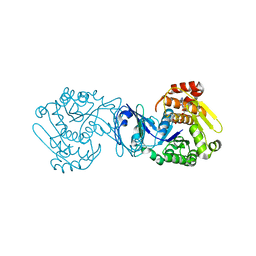 | | Crystal structure of M tuberculosis Adenosine Kinase complexed with AMP-PCP (non-hydrolyzable ATP analog) | | Descriptor: | Adenosine kinase, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Reddy, M.C.M, Palaninathan, S.K, Shetty, N.D, Owen, J.L, Watson, M.D, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-04-18 | | Release date: | 2007-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution crystal structures of Mycobacterium tuberculosis adenosine kinase: insights into the mechanism and specificity of this novel prokaryotic enzyme
J.Biol.Chem., 282, 2007
|
|
1TR7
 
 | | FimH adhesin receptor binding domain from uropathogenic E. coli | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, FimH protein, ... | | Authors: | Bouckaert, J, Berglund, J, Schembri, M, De Genst, E, Cools, L, Wuhrer, M, Hung, C.S, Pinkner, J, Slattegard, R, Zavialov, A, Choudhury, D, Langermann, S, Hultgren, S.J, Wyns, L, Klemm, P, Oscarson, S, Knight, S.D, De Greve, H. | | Deposit date: | 2004-06-21 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Receptor binding studies disclose a novel class of high-affinity inhibitors of the Escherichia coli FimH adhesin
Mol.Microbiol., 55, 2005
|
|
2JQ6
 
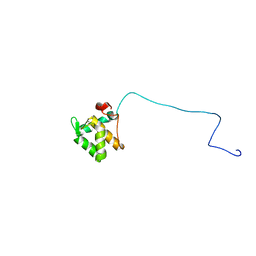 | | Structure of EH-domain of EHD1 | | Descriptor: | CALCIUM ION, EH domain-containing protein 1 | | Authors: | Kieken, F.P, Jovic, M, Caplan, S, Sorgen, P.L. | | Deposit date: | 2007-05-29 | | Release date: | 2008-04-15 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | EH domain of EHD1
J.Biomol.Nmr, 39, 2007
|
|
3N59
 
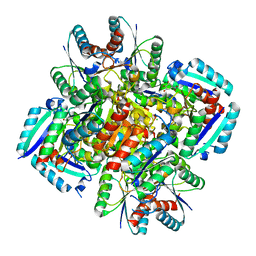 | | Type II dehydroquinase from Mycobacterium Tuberculosis complexed with 3-dehydroshikimate | | Descriptor: | (4S,5R)-4,5-dihydroxy-3-oxocyclohex-1-ene-1-carboxylic acid, 3-dehydroquinate dehydratase, CHLORIDE ION | | Authors: | Snee, W.C, Palaninathan, S.K, Sacchettini, J.C, Dias, M.V.B, Bromfield, K.M, Payne, R, Ciulli, A, Howard, N.I, Abell, C, Blundell, T.L, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-05-24 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural investigation of inhibitor designs targeting 3-dehydroquinate dehydratase from the shikimate pathway of Mycobacterium tuberculosis.
Biochem.J., 436, 2011
|
|
2QHK
 
 | |
