3FCM
 
 | |
3D3A
 
 | |
4JAR
 
 | |
4JAO
 
 | |
4JAT
 
 | |
4JAP
 
 | |
3FJY
 
 | |
3F6A
 
 | |
3SMD
 
 | |
2QVC
 
 | | Crystal structure of a periplasmic sugar ABC transporter from Thermotoga maritima | | Descriptor: | Sugar ABC transporter, periplasmic sugar-binding protein, beta-D-glucopyranose | | Authors: | Palani, K, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-08 | | Release date: | 2007-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a periplasmic glucose-binding protein from Thermotoga maritima.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2QXY
 
 | |
2Y8G
 
 | | Structure of the Ran-binding domain from human RanBP3 (E352A-R353V double mutant) | | Descriptor: | RAN-BINDING PROTEIN 3, SULFATE ION | | Authors: | Langer, K, Dian, C, Rybin, V, Muller, C.W, Petosa, C. | | Deposit date: | 2011-02-06 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Insights Into the Function of the Crm1 Cofactor Ranbp3 from the Structure of its Ran-Binding Domain
Plos One, 6, 2011
|
|
2Y8F
 
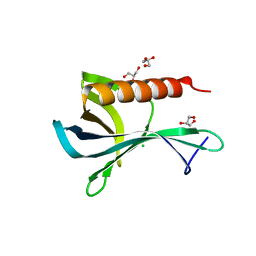 | | Structure of the Ran-binding domain from human RanBP3 (wild type) | | Descriptor: | CHLORIDE ION, GLYCEROL, RAN-BINDING PROTEIN 3 | | Authors: | Langer, K, Dian, C, Rybin, V, Muller, C.W, Petosa, C. | | Deposit date: | 2011-02-06 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights Into the Function of the Crm1 Cofactor Ranbp3 from the Structure of its Ran-Binding Domain
Plos One, 6, 2011
|
|
3BCV
 
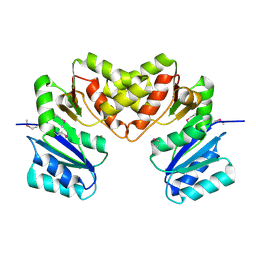 | |
4QRA
 
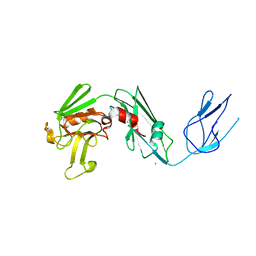 | |
4QRB
 
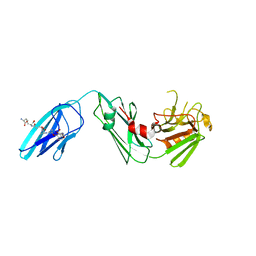 | |
4UU3
 
 | | Ferulic acid decarboxylase from Enterobacter sp. | | Descriptor: | FERULIC ACID DECARBOXYLASE | | Authors: | Hromic, A, Pavkov-Keller, T, Steinkellner, G, Lyskowski, A, Wuensch, C, Gross, J, Fuchs, M, Fauland, K, Glueck, S.M, Faber, K, Gruber, K. | | Deposit date: | 2014-07-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Regioselective Enzymatic Beta-Carboxylation of Para-Hydroxy-Styrene Derivatives Catalyzed by Phenolic Acid Decarboxylases.
Adv. Synth. Catal., 357, 2015
|
|
1I16
 
 | | STRUCTURE OF INTERLEUKIN 16: IMPLICATIONS FOR FUNCTION, NMR, 20 STRUCTURES | | Descriptor: | INTERLEUKIN 16 | | Authors: | Muehlhahn, P, Zweckstetter, M, Georgescu, J, Ciosto, C, Renner, C, Lanzendoerfer, M, Lang, K, Ambrosius, D, Baier, M, Kurth, R, Holak, T.A. | | Deposit date: | 1998-05-20 | | Release date: | 1999-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of interleukin 16 resembles a PDZ domain with an occluded peptide binding site.
Nat.Struct.Biol., 5, 1998
|
|
5ZB1
 
 | | Monomeric crystal structure of orf57 from KSHV | | Descriptor: | ORF57, ZINC ION | | Authors: | Gao, Z.Q, Yuan, F, Lan, K, Dong, Y.H. | | Deposit date: | 2018-02-09 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.061 Å) | | Cite: | The crystal structure of KSHV ORF57 reveals dimeric active sites important for protein stability and function.
PLoS Pathog., 14, 2018
|
|
5ZB3
 
 | | Dimeric crystal structure of ORF57 from KSHV | | Descriptor: | ORF57, ZINC ION | | Authors: | Gao, Z.Q, Yuan, F, Dong, Y.H, Lan, K. | | Deposit date: | 2018-02-09 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.506 Å) | | Cite: | The crystal structure of KSHV ORF57 reveals dimeric active sites important for protein stability and function.
PLoS Pathog., 14, 2018
|
|
4P55
 
 | |
2QLS
 
 | | crystal structure of hemoglobin from dog (Canis familiaris) at 3.5 Angstrom resolution | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Packianathan, C, Sundaresan, S, Palani, K, Neeelagandan, K, Ponnuswamy, M.N. | | Deposit date: | 2007-07-13 | | Release date: | 2008-07-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Purification, Crystallization and Crystal structure analysis of hemoglobin from Dog (Canis familiaris)
To be Published
|
|
4UU2
 
 | | Ferulic acid decarboxylase from Enterobacter sp., single mutant | | Descriptor: | FERULIC ACID DECARBOXYLASE, GLYCINE, PHOSPHATE ION, ... | | Authors: | Hromic, A, Pavkov-Keller, T, Steinkellner, G, Lyskowski, A, Wuensch, C, Gross, J, Fuchs, M, Fauland, K, Glueck, S.M, Faber, K, Gruber, K. | | Deposit date: | 2014-07-24 | | Release date: | 2015-06-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Regioselective Enzymatic Beta-Carboxylation of Para-Hydroxy-Styrene Derivatives Catalyzed by Phenolic Acid Decarboxylases.
Adv. Synth. Catal., 357, 2015
|
|
4V5K
 
 | | Structure of cytotoxic domain of colicin E3 bound to the 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Meenan, N.A.G, Sharma, A, Kelley, A.C, Kleanthous, C, Ramakrishnan, V. | | Deposit date: | 2010-05-29 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis for 16S Ribosomal RNA Cleavage by the Cytotoxic Domain of Colicin E3.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4X1J
 
 | | X-ray crystal structure of the dimeric BMP antagonist NBL1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuroblastoma suppressor of tumorigenicity 1 | | Authors: | Thompson, T.B, Nolan, K, Kattamuri, C. | | Deposit date: | 2014-11-24 | | Release date: | 2015-01-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Neuroblastoma Suppressor of Tumorigenicity 1 (NBL1): INSIGHTS FOR THE FUNCTIONAL VARIABILITY ACROSS BONE MORPHOGENETIC PROTEIN (BMP) ANTAGONISTS.
J.Biol.Chem., 290, 2015
|
|
