3OY8
 
 | |
3OYW
 
 | |
3T2T
 
 | |
3T1L
 
 | |
3T1M
 
 | |
3SIT
 
 | |
3SIS
 
 | |
3PAH
 
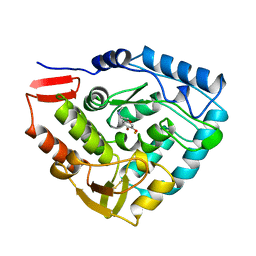 | | HUMAN PHENYLALANINE HYDROXYLASE CATALYTIC DOMAIN DIMER WITH BOUND ADRENALINE INHIBITOR | | Descriptor: | 4-[(1S)-1-hydroxy-2-(methylamino)ethyl]benzene-1,2-diol, FE (III) ION, PHENYLALANINE HYDROXYLASE | | Authors: | Erlandsen, H, Flatmark, T, Stevens, R.C. | | Deposit date: | 1998-08-20 | | Release date: | 1999-04-27 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analysis of the human phenylalanine hydroxylase catalytic domain with bound catechol inhibitors at 2.0 A resolution.
Biochemistry, 37, 1998
|
|
1QDU
 
 | |
4PAH
 
 | |
1TG2
 
 | | Crystal structure of phenylalanine hydroxylase A313T mutant with 7,8-dihydrobiopterin bound | | Descriptor: | 2-AMINO-6-(1,2-DIHYDROXY-PROPYL)-7,8-DIHYDRO-6H-PTERIDIN-4-ONE, FE (III) ION, Phenylalanine-4-hydroxylase | | Authors: | Erlandsen, H, Pey, A.L, Gamez, A, Perez, B, Desviat, L.R, Aguado, C, Koch, R, Surendran, S, Tyring, S, Matalon, R, Scriver, C.R, Ugarte, M, Martinez, A, Stevens, R.C. | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Correction of kinetic and stability defects by tetrahydrobiopterin in phenylketonuria patients with certain phenylalanine hydroxylase mutations.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1TDW
 
 | | Crystal structure of double truncated human phenylalanine hydroxylase BH4-responsive PKU mutant A313T. | | Descriptor: | FE (III) ION, Phenylalanine-4-hydroxylase | | Authors: | Erlandsen, H, Pey, A.L, Gamez, A, Perez, B, Desviat, L.R, Aguado, C, Koch, R, Surendran, S, Tyring, S, Matalon, R, Scriver, C.R, Ugarte, M, Martinez, A, Stevens, R.C. | | Deposit date: | 2004-05-24 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Correction of kinetic and stability defects by tetrahydrobiopterin in phenylketonuria patients with certain phenylalanine hydroxylase mutations.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1F9E
 
 | | CASPASE-8 SPECIFICITY PROBED AT SUBSITE S4: CRYSTAL STRUCTURE OF THE CASPASE-8-Z-DEVD-CHO | | Descriptor: | (PHQ)DEVD, CASPASE-8 ALPHA CHAIN, CASPASE-8 BETA CHAIN | | Authors: | Blanchard, H, Donepudi, M, Tschopp, M, Kodandapani, L, Wu, J.C, Grutter, M.G. | | Deposit date: | 2000-07-10 | | Release date: | 2001-07-10 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Caspase-8 specificity probed at subsite S(4): crystal structure of the caspase-8-Z-DEVD-cho complex.
J.Mol.Biol., 302, 2000
|
|
1DMW
 
 | | CRYSTAL STRUCTURE OF DOUBLE TRUNCATED HUMAN PHENYLALANINE HYDROXYLASE WITH BOUND 7,8-DIHYDRO-L-BIOPTERIN | | Descriptor: | 7,8-DIHYDROBIOPTERIN, FE (III) ION, PHENYLALANINE HYDROXYLASE | | Authors: | Erlandsen, H, Stevens, R.C, Flatmark, T. | | Deposit date: | 1999-12-15 | | Release date: | 2000-03-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and site-specific mutagenesis of pterin-bound human phenylalanine hydroxylase.
Biochemistry, 39, 2000
|
|
7ALO
 
 | | Structure of B*27:09/photoRL9 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Loll, B, Lan, H, Freund, C. | | Deposit date: | 2020-10-07 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exchange catalysis by tapasin exploits conserved and allele-specific features of MHC-I molecules.
Nat Commun, 12, 2021
|
|
8AJ0
 
 | | Mpro of SARS COV-2 in complex with the RK-90 inhibitor | | Descriptor: | (2R,3S)-3-[[(2S)-3-cyclopropyl-2-[2-oxidanylidene-3-(3-phenylpropanoylamino)pyridin-1-yl]propanoyl]amino]-N-methyl-2-oxidanyl-4-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butanamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.519 Å) | | Cite: | Main Protease SARS-COV-2 in complex with the inhibitor RK-90
To Be Published
|
|
8AIQ
 
 | | Mpro of SARS COV-2 in complex with the MG-87 inhibitor | | Descriptor: | CHLORIDE ION, Replicase polyprotein 1ab, ~{tert}-butyl ~{N}-[1-[(2~{S})-1-[[(2~{S},3~{R})-4-azanyl-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-3-cyclopropyl-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Main Protease SARS-COV-2 in complex with the inhibitor MG-87
To Be Published
|
|
8AIV
 
 | | Mpro of SARS COV-2 in complex with the MG-100 inhibitor | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, tert-butyl N-[1-[(2S)-3-cyclopropyl-1-[[(2S,3R)-4-(methylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Main Protease SARS-COV-2 in complex with the inhibitor MG-100
To Be Published
|
|
8AIU
 
 | | Mpro of SARS COV-2 in complex with the MG-97 inhibitor | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION, tert-butyl N-[1-[(2S)-3-cyclopropyl-1-[[(2S,3R)-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3S)-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Main Protease SARS-COV-2 in complex with the inhibitor MG-97
To Be Published
|
|
8AIZ
 
 | | Mpro of SARS-CoV-2 in complex with the RK-68 inhibitor | | Descriptor: | (2~{R},3~{S})-3-[[(2~{S})-3-cyclopropyl-2-[2-oxidanylidene-3-(2-phenylethanoylamino)pyridin-1-yl]propanoyl]amino]-~{N}-methyl-2-oxidanyl-4-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butanamide, CHLORIDE ION, Replicase polyprotein 1ab | | Authors: | El Kilani, H, Hilgenfeld, R. | | Deposit date: | 2022-07-27 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Main Protease SARS-CoV-2 in complex with the inhibitor RK-68
To Be Published
|
|
3TAY
 
 | |
3TB0
 
 | |
1AJ5
 
 | | CALPAIN DOMAIN VI APO | | Descriptor: | CALPAIN | | Authors: | Cygler, M, Grochulski, P, Blanchard, H. | | Deposit date: | 1997-05-15 | | Release date: | 1998-05-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a calpain Ca(2+)-binding domain reveals a novel EF-hand and Ca(2+)-induced conformational changes.
Nat.Struct.Biol., 4, 1997
|
|
4UG1
 
 | | GpsB N-terminal domain | | Descriptor: | CELL CYCLE PROTEIN GPSB, IMIDAZOLE, NICKEL (II) ION | | Authors: | Rismondo, J, Cleverley, R.M, Lane, H.V, Grohennig, S, Steglich, A, Muller, L, Krishna Mannala, G, Hain, T, Lewis, R.J, Halbedel, S. | | Deposit date: | 2015-03-20 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Bacterial Cell Division Determinant Gpsb and its Interaction with Penicillin Binding Proteins.
Mol.Microbiol., 99, 2016
|
|
6UGT
 
 | | Crystal structure of the Fab fragment of PF06438179/GP1111 an infliximab biosimilar in a I-centered orthorhombic crystal form, Lot A | | Descriptor: | 1,2-ETHANEDIOL, PF06438179 Fab Heavy Chain, PF06438179 Fab Light Chain | | Authors: | Lerch, T.F, Sharpe, P, Mayclin, S.J, Edwards, T.E, Polleck, S, Rouse, J.C, Conlan, H.D. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
BioDrugs, 34, 2020
|
|
