8ASM
 
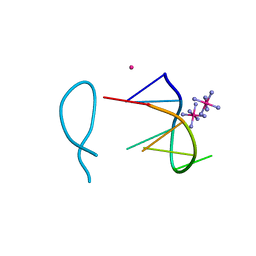 | | Cobalt(II) bound to a non-canonical quadruplex | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), DNA (5'-D(*GP*CP*AP*TP*GP*CP*T)-3') | | Authors: | Lambert, M.C, Hall, J.P. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identifying metal-DNA binding sites, what is the best method to get transition metals into a crystal system?
To Be Published
|
|
8ASO
 
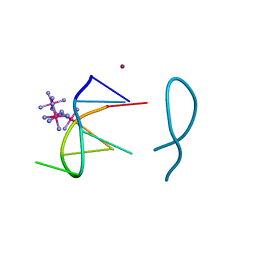 | | Nickel(II) bound to a non-canonical quadruplex | | Descriptor: | COBALT HEXAMMINE(III), DNA (5'-D(*GP*CP*AP*TP*GP*CP*T)-3'), NICKEL (II) ION | | Authors: | Lambert, M.C, Hall, J.P. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Identifying metal-DNA binding sites, what is the best method to get transition metals into a crystal system?
To Be Published
|
|
3DY6
 
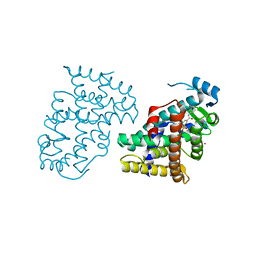 | | PPARdelta complexed with an anthranilic acid partial agonist | | Descriptor: | 2-({[3-(3,4-dihydroisoquinolin-2(1H)-ylsulfonyl)phenyl]carbonyl}amino)benzoic acid, IODIDE ION, Peroxisome proliferator-activated receptor delta | | Authors: | Lambert, M.L, Xu, R, Shearer, B.G, Wilson, T.M. | | Deposit date: | 2008-07-25 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a novel class of PPARdelta partial agonists
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3S9S
 
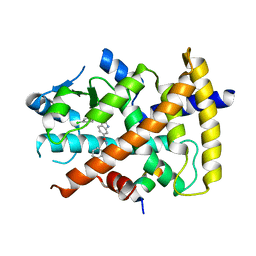 | | Ligand binding domain of PPARgamma complexed with a benzimidazole partial agonist | | Descriptor: | 1-(3,4-dichlorobenzyl)-2-methyl-N-[(1R)-1-phenylpropyl]-1H-benzimidazole-5-carboxamide, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Lambert, M.H, Xu, R.X. | | Deposit date: | 2011-06-02 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of GSK1997132B a novel centrally penetrant benzimidazole PPARgamma partial agonist.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3PEQ
 
 | | PPARd complexed with a phenoxyacetic acid partial agonist | | Descriptor: | IODIDE ION, Peroxisome proliferator-activated receptor delta, [(4-{butyl[2-methyl-4'-(methylsulfanyl)biphenyl-3-yl]sulfamoyl}naphthalen-1-yl)oxy]acetic acid, ... | | Authors: | Lambert, M.H, Evans, K.A, Shearer, B.G, Wisnoski, D.D, Shi, D, Jin, J, Rivero, R.A, Sparks, S.M, Winegar, D.A, Billin, A.N, Britt, C, Way, J.M, Leesnitzer, L.M, Merrihew, R.V. | | Deposit date: | 2010-10-27 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phenoxyacetic acid PPARd partial agonists for the treatment of type 2 diabetes: synthesis, optimization, and in vivo efficacy
Bioorg.Med.Chem.Lett., 21, 2011
|
|
8BAE
 
 | |
8BAF
 
 | |
8BAG
 
 | |
8OEA
 
 | |
8OE7
 
 | |
8OE8
 
 | |
8OE9
 
 | |
8OEZ
 
 | |
8OEB
 
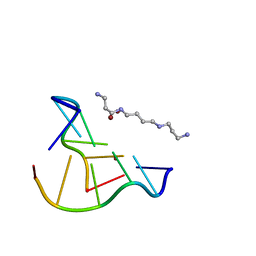 | |
8OE3
 
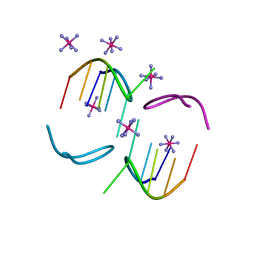 | |
8OEC
 
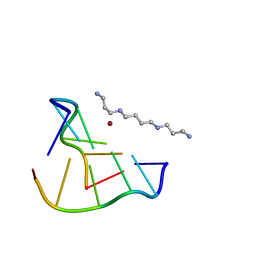 | |
8OEX
 
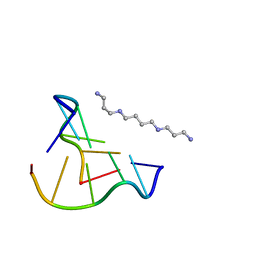 | |
8Y4Z
 
 | | Monomeric HERC5 HECT c-lobe structure in solution | | Descriptor: | E3 ISG15--protein ligase HERC5 | | Authors: | Dag, C, Lambert, M, Kahraman, K, Lohn, F, Lee, W, Gocenler, O, Guntert, P, Dotsch, V. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Monomeric HERC5 HECT c-lobe structure in solution
To Be Published
|
|
7AMA
 
 | | IL-17A in complex with small molecule modulators | | Descriptor: | Interleukin-17A, ~{N}-[(2~{S})-1,1-dicyclopropyl-3-[[4-(3,5-dimethyl-1~{H}-pyrazol-4-yl)phenyl]amino]-3-oxidanylidene-propan-2-yl]-2-propan-2-yl-pyrazole-3-carboxamide | | Authors: | Hakansson, M, Kimbung, R, Logan, D, Walse, U.B, de Groot, M.J, Andrews, M.D, Dack, K.N, Lambert, M. | | Deposit date: | 2020-10-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of an Oral, Rule of 5 Compliant, Interleukin 17A Protein-Protein Interaction Modulator for the Potential Treatment of Psoriasis and Other Inflammatory Diseases.
J.Med.Chem., 65, 2022
|
|
7AMG
 
 | | IL-17A in complex with small molecule modulators | | Descriptor: | (3~{R})-4-[4-[[(2~{S})-2-[[2,2-bis(fluoranyl)-2-phenyl-ethanoyl]amino]-3-(2-chlorophenyl)propanoyl]amino]phenyl]-3-[[(2~{S})-3-methyl-2-[2-[2-[(2-methylpropan-2-yl)oxycarbonylamino]ethoxy]ethanoylamino]butanoyl]amino]butanoic acid, Interleukin-17A | | Authors: | Hakansson, M, Kimbung, R, Logan, D, Walse, U.B, de Groot, M.J, Andrews, M.D, Dack, K.N, Lambert, M. | | Deposit date: | 2020-10-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Discovery of an Oral, Rule of 5 Compliant, Interleukin 17A Protein-Protein Interaction Modulator for the Potential Treatment of Psoriasis and Other Inflammatory Diseases.
J.Med.Chem., 65, 2022
|
|
1CGL
 
 | | Structure of the catalytic domain of fibroblast collagenase complexed with an inhibitor | | Descriptor: | CALCIUM ION, FIBROBLAST COLLAGENASE, N-[(1S)-3-{[(benzyloxy)carbonyl]amino}-1-carboxypropyl]-L-leucyl-N-(2-morpholin-4-ylethyl)-L-phenylalaninamide, ... | | Authors: | Lovejoy, B, Cleasby, A, Hassell, A.M, Longley, K, Luther, M.A, Weigl, D, Mcgeehan, G, Mcelroy, A.B, Drewry, D, Lambert, M.H, Jordan, S.R. | | Deposit date: | 1993-11-17 | | Release date: | 1995-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the catalytic domain of fibroblast collagenase complexed with an inhibitor.
Science, 263, 1994
|
|
1KKQ
 
 | | Crystal structure of the human PPAR-alpha ligand-binding domain in complex with an antagonist GW6471 and a SMRT corepressor motif | | Descriptor: | N-((2S)-2-({(1Z)-1-METHYL-3-OXO-3-[4-(TRIFLUOROMETHYL) PHENYL]PROP-1-ENYL}AMINO)-3-{4-[2-(5-METHYL-2-PHENYL-1,3-OXAZOL-4-YL)ETHOXY]PHENYL}PROPYL)PROPANAMIDE, NUCLEAR RECEPTOR CO-REPRESSOR 2, PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR | | Authors: | Xu, H.E, Stanley, T.B, Montana, V.G, Lambert, M.H, Shearer, B.G, Cobb, J.E, McKee, D.D, Galardi, C.M, Nolte, R.T, Parks, D.J. | | Deposit date: | 2001-12-10 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha.
Nature, 415, 2002
|
|
1G1U
 
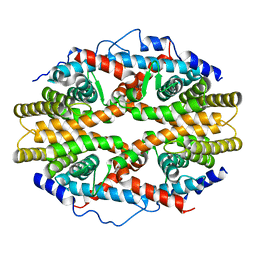 | | THE 2.5 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE RXRALPHA LIGAND BINDING DOMAIN IN TETRAMER IN THE ABSENCE OF LIGAND | | Descriptor: | RETINOIC ACID RECEPTOR RXR-ALPHA | | Authors: | Gampe Jr, R.T, Montana, V.G, Lambert, M.H, Wisely, G.B, Milburn, M.V, Xu, H.E. | | Deposit date: | 2000-10-13 | | Release date: | 2001-04-25 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for autorepression of retinoid X receptor by tetramer formation and the AF-2 helix.
Genes Dev., 14, 2000
|
|
1G5Y
 
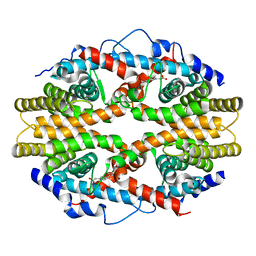 | | THE 2.0 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF THE RXRALPHA LIGAND BINDING DOMAIN TETRAMER IN THE PRESENCE OF A NON-ACTIVATING RETINOIC ACID ISOMER. | | Descriptor: | RETINOIC ACID, RETINOIC ACID RECEPTOR RXR-ALPHA | | Authors: | Gampe Jr, R.T, Montana, V.G, Lambert, M.H, Wisely, G.B, Milburn, M.V, Xu, H.E. | | Deposit date: | 2000-11-02 | | Release date: | 2001-05-02 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for autorepression of retinoid X receptor by tetramer formation and the AF-2 helix.
Genes Dev., 14, 2000
|
|
2P54
 
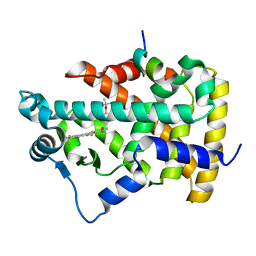 | | a crystal structure of PPAR alpha bound with SRC1 peptide and GW735 | | Descriptor: | 2-METHYL-2-(4-{[({4-METHYL-2-[4-(TRIFLUOROMETHYL)PHENYL]-1,3-THIAZOL-5-YL}CARBONYL)AMINO]METHYL}PHENOXY)PROPANOIC ACID, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor alpha | | Authors: | Xu, R.X, Xu, H.E, Sierra, M.L, Montana, V.G, Lambert, M.H, Pianetti, P.M. | | Deposit date: | 2007-03-14 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Substituted 2-[(4-Aminomethyl)phenoxy]-2-methylpropionic Acid PPAR Agonists. 1.Discovery of a Novel Series of Potent HDLc Raising Agents.
J.Med.Chem., 50, 2007
|
|
