7UHO
 
 | | Time-Resolved Structure of Metallo Beta-Lactamase L1 in a Complex with Cleaved Moxalactam (500 ms Snapshot) | | Descriptor: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Srajer, V, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-03-27 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
1NG9
 
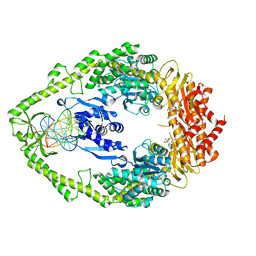 | | E.coli MutS R697A: an ATPase-asymmetry mutant | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP*GP*AP*CP*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lamers, M.H, Winterwerp, H.H.K, Sixma, T.K. | | Deposit date: | 2002-12-17 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The alternating ATPase domains of MutS control DNA mismatch repair
Embo J., 22, 2003
|
|
7JHE
 
 | | Room Temperature Structure of SARS-CoV-2 Nsp10/Nsp16 Methyltransferase in a Complex with 2'-O-methylated m7GpppA Cap-1 and SAH Determined by Fixed-Target Serial Crystallography | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | Authors: | Wilamowski, M, Sherrell, D.A, Minasov, G, Kim, Y, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-20 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4DG4
 
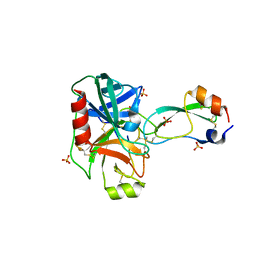 | | Human mesotrypsin-S39Y complexed with bovine pancreatic trypsin inhibitor (BPTI) | | Descriptor: | CALCIUM ION, PRSS3 protein, Pancreatic trypsin inhibitor, ... | | Authors: | Salameh, M.A, Soares, A.S, Radisky, E.S. | | Deposit date: | 2012-01-24 | | Release date: | 2012-09-12 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Presence versus absence of hydrogen bond donor Tyr-39 influences interactions of cationic trypsin and mesotrypsin with protein protease inhibitors.
Protein Sci., 21, 2012
|
|
8OE8
 
 | |
8OE7
 
 | |
8OEA
 
 | |
3AUH
 
 | | A simplified BPTI variant with poly Arg amino acid tag (C3R) at the C-terminus | | Descriptor: | Bovine pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M, Kato, A, Khan, M.M.A, Noguchi, K, Yohda, M, Kidokoro, S.I, Kuroda, Y. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Effect of amino acid mutations on protein's solubility, function and structure characterized using short poly amino acid peptide tags
To be Published
|
|
3AUE
 
 | | A simplified BPTI variant with poly His amino acid tag (C5H) at the C-terminus | | Descriptor: | Bovine pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M, Kato, A, Khan, M.M.A, Noguchi, K, Yohda, M, Kidokoro, S.I, Kuroda, Y. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Effect of amino acid mutations on protein's solubility, function and structure characterized using short poly amino acid peptide tags
To be Published
|
|
7VOI
 
 | | Structure of the human CNOT1(MIF4G)-CNOT6L-CNOT7 complex | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 6-like, CCR4-NOT transcription complex subunit 7 | | Authors: | Bartlam, M, Zhang, Q. | | Deposit date: | 2021-10-13 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.38 Å) | | Cite: | Structure of the human Ccr4-Not nuclease module using X-ray crystallography and electron paramagnetic resonance spectroscopy distance measurements.
Protein Sci., 31, 2022
|
|
8BAG
 
 | |
8BAE
 
 | |
8BAF
 
 | |
7OTO
 
 | | The structure of MutS bound to two molecules of AMPPNP | | Descriptor: | DNA mismatch repair protein MutS, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Lamers, M.H, Borsellini, A, Friedhoff, P, Kunetsky, V. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryogenic electron microscopy structures reveal how ATP and DNA binding in MutS coordinates sequential steps of DNA mismatch repair.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7OU4
 
 | | The structure of MutS bound to one molecule of ATP and one molecule of ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, DNA mismatch repair protein MutS, ... | | Authors: | Lamers, M.H, Borsellini, A, Friedhoff, P, Kunetsky, V. | | Deposit date: | 2021-06-11 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryogenic electron microscopy structures reveal how ATP and DNA binding in MutS coordinates sequential steps of DNA mismatch repair.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7OU0
 
 | | The structure of MutS bound to two molecules of ADP-Vanadate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein MutS, MAGNESIUM ION, ... | | Authors: | Lamers, M.H, Borsellini, A, Friedhoff, P, Kunetsky, V. | | Deposit date: | 2021-06-10 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryogenic electron microscopy structures reveal how ATP and DNA binding in MutS coordinates sequential steps of DNA mismatch repair.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7OU2
 
 | | The structure of MutS bound to two molecules of ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein MutS | | Authors: | Lamers, M.H, Borsellini, A, Friedhoff, P, Kunetsky, V. | | Deposit date: | 2021-06-11 | | Release date: | 2022-01-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryogenic electron microscopy structures reveal how ATP and DNA binding in MutS coordinates sequential steps of DNA mismatch repair.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3PEQ
 
 | | PPARd complexed with a phenoxyacetic acid partial agonist | | Descriptor: | IODIDE ION, Peroxisome proliferator-activated receptor delta, [(4-{butyl[2-methyl-4'-(methylsulfanyl)biphenyl-3-yl]sulfamoyl}naphthalen-1-yl)oxy]acetic acid, ... | | Authors: | Lambert, M.H, Evans, K.A, Shearer, B.G, Wisnoski, D.D, Shi, D, Jin, J, Rivero, R.A, Sparks, S.M, Winegar, D.A, Billin, A.N, Britt, C, Way, J.M, Leesnitzer, L.M, Merrihew, R.V. | | Deposit date: | 2010-10-27 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phenoxyacetic acid PPARd partial agonists for the treatment of type 2 diabetes: synthesis, optimization, and in vivo efficacy
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3AUG
 
 | | A simplified BPTI variant with poly Pro amino acid tag (C5P) at the C-terminus | | Descriptor: | Bovine pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M, Kato, A, Khan, M.M.A, Noguchi, K, Yohda, M, Kidokoro, S.I, Kuroda, Y. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Effect of amino acid mutations of protein's solubility, function and structure characterized using short poly amino acid peptide tags
To be Published
|
|
3AUD
 
 | | Simplified BPTI variant with poly Asn amino acid tag (C5N) at the C-terminus | | Descriptor: | Bovine pancreatic trypsin inhibitor | | Authors: | Islam, M.M, Kato, A, Khan, M.M.A, Noguchi, K, Yohda, M, Kidokoro, S.I, Kuroda, Y. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | Effect of amino acid mutations on protein's solubility, function and structure characterized using short poly amino acid peptide tags
To be Published
|
|
3AUB
 
 | | A simplified BPTI variant stabilized by the A14G and A38V substitutions | | Descriptor: | Bovine Pancreatic trypsin inhibitor | | Authors: | Islam, M.M, Kato, A, Khan, M.M.A, Noguchi, K, Yohda, M, Kidokoro, S.I, Kuroda, Y. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Effect of amino acid mutations of protein's solubility, function and structure characterized using short poly amino acid peptide tags
To be Published
|
|
3AUI
 
 | | A simplified BPTI variant with poly Glu amino acid tag (C3E) at the C-terminus | | Descriptor: | Bovine pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M, Kato, A, Khan, M.M.A, Noguchi, K, Yohda, M, Kidokoro, S.I, Kuroda, Y. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Effect of amino acid mutations on protein's solubility, function and structure characterized using short poly amino acid peptide tags
To be Published
|
|
3AUC
 
 | | A simplified BPTI variant with poly SER (C5S) amino acid tag at the c-terminus | | Descriptor: | Bovine pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Islam, M.M, Kato, A, Khan, M.M.A, Noguchi, K, Yohda, M, Kidokoro, S.I, Kuroda, Y. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Effect of amino acid mutations on protein's solubility, function and structure characterized using short poly amino acid peptide tags
To be Published
|
|
8OE3
 
 | |
8OEB
 
 | |
