7YZO
 
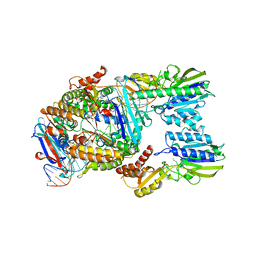 | | Endonuclease state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (31-MER), MAGNESIUM ION, ... | | Authors: | Gut, F, Kaeshammer, L, Lammens, K, Bartho, J, van de Logt, E, Kessler, B, Hopfner, K.P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50.
Mol.Cell, 82, 2022
|
|
7Z03
 
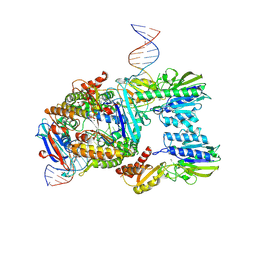 | | Endonuclease state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ADP and extended dsDNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (39-MER), MAGNESIUM ION, ... | | Authors: | Gut, F, Kaeshammer, L, Lammens, K, Bartho, J, van de Logt, E, Kessler, B, Hopfner, K.P. | | Deposit date: | 2022-02-21 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50.
Mol.Cell, 82, 2022
|
|
5DAC
 
 | | ATP-gamma-S bound Rad50 from Chaetomium thermophilum in complex with DNA | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3'), ... | | Authors: | Seifert, F.U, Lammens, K, Stoehr, G, Kessler, B, Hopfner, K.-P. | | Deposit date: | 2015-08-19 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50.
Embo J., 35, 2016
|
|
5DA9
 
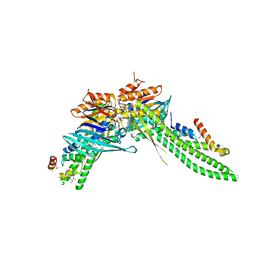 | | ATP-gamma-S bound Rad50 from Chaetomium thermophilum in complex with the Rad50-binding domain of Mre11 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Putative double-strand break protein, ... | | Authors: | Seifert, F.U, Lammens, K, Stoehr, G, Kessler, B, Hopfner, K.-P. | | Deposit date: | 2015-08-19 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural mechanism of ATP-dependent DNA binding and DNA end bridging by eukaryotic Rad50.
Embo J., 35, 2016
|
|
7ZKE
 
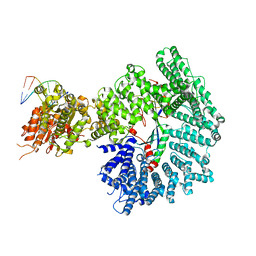 | | Mot1:TBP:DNA - pre-hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (36-MER), ... | | Authors: | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6S6V
 
 | | Resting state of the E. coli Mre11-Rad50 (SbcCD) head complex bound to ATPgS | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Nuclease SbcCD subunit C, ... | | Authors: | Kaeshammer, L, Saathoff, J.H, Gut, F, Bartho, J, Alt, A, Kessler, B, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-07-03 | | Release date: | 2019-09-04 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of DNA End Sensing and Processing by the Mre11-Rad50 Complex.
Mol.Cell, 76, 2019
|
|
7ZES
 
 | | Human SLFN11 dimer bound to ssDNA | | Descriptor: | DNA (5'-D(P*CP*GP*CP*GP*T)-3'), MAGNESIUM ION, Schlafen family member 11, ... | | Authors: | Metzner, F.J, Kugler, M, Wenzl, S.J, Lammens, K. | | Deposit date: | 2022-03-31 | | Release date: | 2022-09-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanistic understanding of human SLFN11.
Nat Commun, 13, 2022
|
|
7ZEL
 
 | | Human SLFN11 dimer apoenzyme | | Descriptor: | MAGNESIUM ION, Schlafen family member 11, ZINC ION | | Authors: | Metzner, F.J, Kugler, M, Wenzl, S.J, Lammens, K. | | Deposit date: | 2022-03-31 | | Release date: | 2022-09-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanistic understanding of human SLFN11.
Nat Commun, 13, 2022
|
|
7ZEP
 
 | | Human SLFN11 E209A dimer | | Descriptor: | Schlafen family member 11, ZINC ION | | Authors: | Metzner, F.J, Kugler, M, Wenzl, S.J, Lammens, K. | | Deposit date: | 2022-03-31 | | Release date: | 2022-09-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanistic understanding of human SLFN11.
Nat Commun, 13, 2022
|
|
7Z8S
 
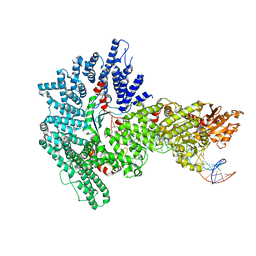 | | Mot1:TBP:DNA - post hydrolysis state | | Descriptor: | DNA (36-MER), Helicase-like protein, Putative tata-box binding protein | | Authors: | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | Deposit date: | 2022-03-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z7N
 
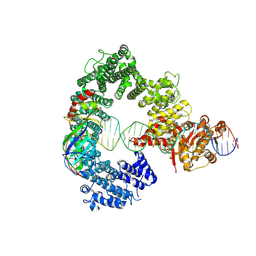 | | Mot1E1434Q:TBP:DNA - substrate recognition state | | Descriptor: | DNA (36-MER), Helicase-like protein, Putative tata-box binding protein | | Authors: | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZB5
 
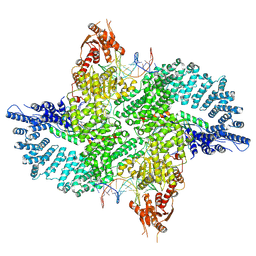 | | Mot1(1-1836):TBP:DNA - post-hydrolysis complex dimer | | Descriptor: | DNA (36-MER), Helicase-like protein, Putative tata-box binding protein | | Authors: | Woike, S, Eustermann, S, Jung, J, Wenzl, S.J, Hagemann, G, Bartho, J.D, Lammens, K, Butryn, A, Herzog, F, Hopfner, K.-P. | | Deposit date: | 2022-03-23 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for TBP displacement from TATA box DNA by the Swi2/Snf2 ATPase Mot1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2R8J
 
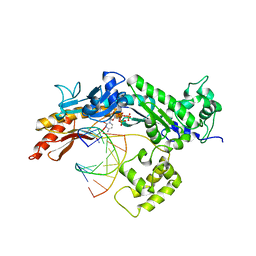 | | Structure of the Eukaryotic DNA Polymerase eta in complex with 1,2-d(GpG)-cisplatin containing DNA | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, Cisplatin, ... | | Authors: | Carell, T, Alt, A, Lammens, K. | | Deposit date: | 2007-09-11 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta
Science, 318, 2007
|
|
2R8K
 
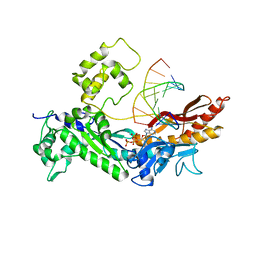 | | Structure of the Eukaryotic DNA Polymerase eta in complex with 1,2-d(GpG)-cisplatin containing DNA | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*DGP*DTP*DGP*DGP*DTP*DGP*DAP*DGP*DC)-3', 5'-D(P*DGP*DGP*DCP*DTP*DCP*DAP*DCP*DCP*DAP*DC)-3', ... | | Authors: | Carell, T, Alt, A, Lammens, K. | | Deposit date: | 2007-09-11 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta
Science, 318, 2007
|
|
3THN
 
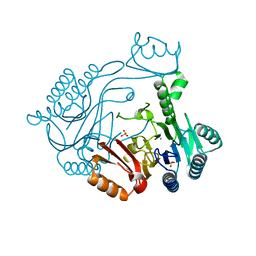 | | Crystal structure of Mre11 core with manganese | | Descriptor: | Exonuclease, putative, MANGANESE (II) ION, ... | | Authors: | Moeckel, C, Lammens, K. | | Deposit date: | 2011-08-19 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.811 Å) | | Cite: | ATP driven structural changes of the bacterial Mre11:Rad50 catalytic head complex.
Nucleic Acids Res., 40, 2012
|
|
3THO
 
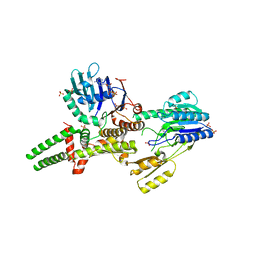 | |
7RJO
 
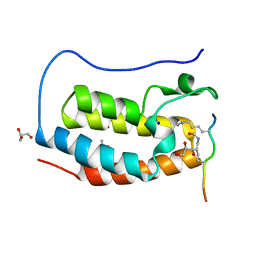 | | Crystal structure of human Bromodomain containing protein 4 (BRD4) in complex with hnRNPK | | Descriptor: | ACETATE ION, Bromodomain-containing protein 4, GLYCEROL, ... | | Authors: | Fedorov, E, Islam, K, Ghosh, A. | | Deposit date: | 2021-07-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Uncovering the Bromodomain Interactome using Site-Specific Azide-Acetyllysine Photochemistry, Proteomic Profiling and Structural Characterization
Biorxiv, 2021
|
|
7RJQ
 
 | | Crystal structure of human Bromodomain containing protein 4 (BRD4) in complex with ILF3 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, CHLORIDE ION, ... | | Authors: | Fedorov, E, Islam, K, Ghosh, A. | | Deposit date: | 2021-07-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Uncovering the Bromodomain Interactome using Site-Specific Azide-Acetyllysine Photochemistry, Proteomic Profiling and Structural Characterization
To Be Published
|
|
7RJR
 
 | | Crystal structure of human Bromodomain containing protein 4 (BRD4) in complex with BCLTF1 | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2-associated transcription factor 1, Bromodomain-containing protein 4, ... | | Authors: | Fedorov, E, Islam, K, Ghosh, A. | | Deposit date: | 2021-07-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Uncovering the Bromodomain Interactome using Site-Specific Azide-Acetyllysine Photochemistry, Proteomic Profiling and Structural Characterization
To Be Published
|
|
7RJL
 
 | | Crystal structure of human Bromodomain containing protein 3 (BRD3) in complex with SHMT | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, GLYCEROL, ... | | Authors: | Fedorov, E, Islam, K, Ghosh, A. | | Deposit date: | 2021-07-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Uncovering the Bromodomain Interactome using Site-Specific Azide-Acetyllysine Photochemistry, Proteomic Profiling and Structural Characterization
Biorxiv, 2021
|
|
7RJM
 
 | | Crystal structure of human Bromodomain containing protein 3 (BRD3) in complex with ILF3 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, Interleukin enhancer-binding factor 3 | | Authors: | Fedorov, E, Islam, K, Ghosh, A. | | Deposit date: | 2021-07-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Uncovering the Bromodomain Interactome using Site-Specific Azide-Acetyllysine Photochemistry, Proteomic Profiling and Structural Characterization
Biorxiv, 2021
|
|
7RJK
 
 | | Crystal structure of human Bromodomain containing protein 3 (BRD3) in complex with hnRNPK | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, GLYCEROL, ... | | Authors: | Fedorov, E, Islam, K, Ghosh, A. | | Deposit date: | 2021-07-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Uncovering the Bromodomain Interactome using Site-Specific Azide-Acetyllysine Photochemistry, Proteomic Profiling and Structural Characterization
Biorxiv, 2021
|
|
7RJP
 
 | | Crystal structure of human Bromodomain containing protein 4 (BRD4) in complex with SHMT | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, CHLORIDE ION, ... | | Authors: | Fedorov, E, Islam, K, Ghosh, A. | | Deposit date: | 2021-07-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Uncovering the Bromodomain Interactome using Site-Specific Azide-Acetyllysine Photochemistry, Proteomic Profiling and Structural Characterization
To Be Published
|
|
7RJN
 
 | | Crystal structure of human bromodomain containing protein 3 (BRD3) in complex with BCLTF1 | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2-associated transcription factor 1, Bromodomain-containing protein 3 | | Authors: | Fedorov, E, Islam, K, Ghosh, A. | | Deposit date: | 2021-07-21 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Uncovering the Bromodomain Interactome using Site-Specific Azide-Acetyllysine Photochemistry, Proteomic Profiling and Structural Characterization
Biorxiv, 2021
|
|
4FBK
 
 | | Crystal structure of a covalently fused Nbs1-Mre11 complex with one manganese ion per active site | | Descriptor: | DNA repair and telomere maintenance protein nbs1,DNA repair protein rad32 CHIMERIC PROTEIN, MANGANESE (II) ION, SULFATE ION | | Authors: | Schiller, C.B, Lammens, K, Hopfner, K.P. | | Deposit date: | 2012-05-23 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.379 Å) | | Cite: | Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling.
Nat.Struct.Mol.Biol., 19, 2012
|
|
