6MHJ
 
 | | Structure of BoNT mutant | | Descriptor: | Botulinum neurotoxin type A, PHOSPHATE ION | | Authors: | Lam, K, Jin, R. | | Deposit date: | 2018-09-18 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.019 Å) | | Cite: | A viral-fusion-peptide-like molecular switch drives membrane insertion of botulinum neurotoxin A1.
Nat Commun, 9, 2018
|
|
4FQ0
 
 | |
4GC8
 
 | |
5WIX
 
 | | Crystal structure of P47 of Clostridium botulinum E1 | | Descriptor: | p-47 protein | | Authors: | Lam, K, Liu, S, Qi, R, Jin, R. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The hypothetical protein P47 of Clostridium botulinum E1 strain Beluga has a structural topology similar to bactericidal/permeability-increasing protein.
Toxicon, 147, 2018
|
|
7NA9
 
 | | Crystal structure of BoNT/B-LC-JSG-C1 | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin type B, JSG-C1, ... | | Authors: | Lam, K, Jin, R. | | Deposit date: | 2021-06-20 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Probing the structure and function of the protease domain of botulinum neurotoxins using single-domain antibodies.
Plos Pathog., 18, 2022
|
|
7L6V
 
 | |
7M1H
 
 | |
7LZP
 
 | | LC/A-JPU-B9-JPU-A11-JPU-G11 | | Descriptor: | Botulinum neurotoxin A light chain, JPU-A11, JPU-B9, ... | | Authors: | Lam, K, Jin, R. | | Deposit date: | 2021-03-10 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Probing the structure and function of the protease domain of botulinum neurotoxins using single-domain antibodies.
Plos Pathog., 18, 2022
|
|
7K84
 
 | |
7K7Y
 
 | |
6UFT
 
 | |
8FBF
 
 | | Crystal structure of OrfX2 from Clostridium botulinum E1 | | Descriptor: | Neurotoxin complex component Orf-X2, SULFATE ION | | Authors: | Lam, K.H, Gao, L. | | Deposit date: | 2022-11-29 | | Release date: | 2022-12-21 | | Last modified: | 2023-03-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of OrfX1, OrfX2 and the OrfX1-OrfX3 complex from the orfX gene cluster of botulinum neurotoxin E1.
Febs Lett., 597, 2023
|
|
6UL6
 
 | |
6UC6
 
 | |
6UI1
 
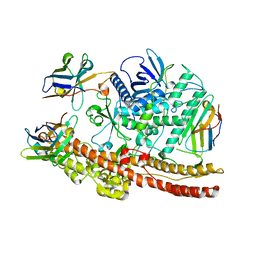 | | Crystal structure of BoNT/A-LCHn domain in complex with VHH ciA-D12, ciA-B5, and ciA-H7 | | Descriptor: | BoNT/A, ciA-B5, ciA-D12, ... | | Authors: | Lam, K, Jin, R. | | Deposit date: | 2019-09-29 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.20000863 Å) | | Cite: | Structural Insights into Rational Design of Single-Domain Antibody-Based Antitoxins against Botulinum Neurotoxins
Cell Rep, 30, 2020
|
|
6UHT
 
 | |
6UL4
 
 | |
3USY
 
 | |
3GWG
 
 | |
3H1G
 
 | |
3H1F
 
 | |
3H1E
 
 | | Crystal structure of Mg(2+) and BeH(3)(-)-bound CheY of Helicobacter pylori | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein cheY homolog, MAGNESIUM ION, ... | | Authors: | Lam, K.H, Ling, T.K, Au, S.W. | | Deposit date: | 2009-04-12 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of activated CheY1 from Helicobacter pylori.
J.Bacteriol., 192, 2010
|
|
6DKK
 
 | | Structure of BoNT | | Descriptor: | Botulinum neurotoxin type A, PHOSPHATE ION | | Authors: | Lam, K, Jin, R. | | Deposit date: | 2018-05-29 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A viral-fusion-peptide-like molecular switch drives membrane insertion of botulinum neurotoxin A1.
Nat Commun, 9, 2018
|
|
4UJA
 
 | | Protein Kinase A in complex with an Inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-{(3S,4R)-4-[(5-bromothiophen-2-yl)carbonyl]pyrrolidin-3-yl}quinazolin-4(3H)-one, BROMIDE ION, ... | | Authors: | Alam, K.A, Engh, R.A. | | Deposit date: | 2015-04-09 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Addressing the Glycine-Rich Loop of Protein Kinases by a Multi-Facetted Interaction Network: Inhibition of Pka and a Pkb Mimic.
Chemistry, 22, 2016
|
|
4UJ2
 
 | | Protein Kinase A in complex with an Inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-[(3S,4R)-4-(3-iodanylphenyl)carbonylpyrrolidin-3-yl]-3H-quinazolin-4-one, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, ... | | Authors: | Alam, K.A, Engh, R.A. | | Deposit date: | 2015-04-07 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.019 Å) | | Cite: | Addressing the Glycine-Rich Loop of Protein Kinases by a Multi-Facetted Interaction Network: Inhibition of Pka and a Pkb Mimic.
Chemistry, 22, 2016
|
|
