2VPM
 
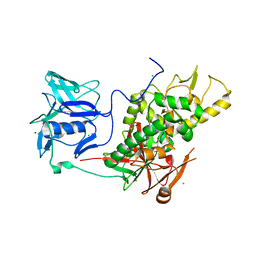 | | Trypanothione synthetase | | Descriptor: | BROMIDE ION, CHLORIDE ION, TRYPANOTHIONE SYNTHETASE | | Authors: | Fyfe, P.K, Oza, S.L, Fairlamb, A.H, Hunter, W.N. | | Deposit date: | 2008-03-03 | | Release date: | 2008-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Leishmania Trypanothione Synthetase-Amidase Structure Reveals a Basis for Regulation of Conflicting Synthetic and Hydrolytic Activities.
J.Biol.Chem., 283, 2008
|
|
3HGW
 
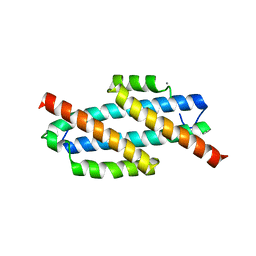 | | Apo Structure of Pseudomonas aeruginosa Isochorismate-Pyruvate Lyase I87T mutant | | Descriptor: | CALCIUM ION, Salicylate biosynthesis protein pchB | | Authors: | Luo, Q, Lamb, A.L. | | Deposit date: | 2009-05-14 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-function analyses of isochorismate-pyruvate lyase from Pseudomonas aeruginosa suggest differing catalytic mechanisms for the two pericyclic reactions of this bifunctional enzyme
Biochemistry, 48, 2009
|
|
3HGX
 
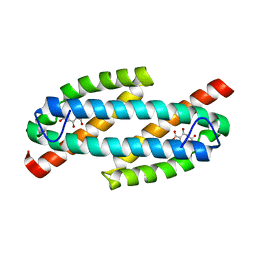 | |
3HHP
 
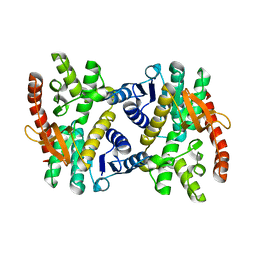 | |
3SQW
 
 | |
3REM
 
 | | Structure of the Isochorismate-Pyruvate Lyase from Pseudomonas aerugionsa with Bound Salicylate and Pyruvate | | Descriptor: | 2-HYDROXYBENZOIC ACID, PYRUVIC ACID, Salicylate biosynthesis protein pchB | | Authors: | Olucha, J, Ouellette, A.N, Luo, Q, Lamb, A.L. | | Deposit date: | 2011-04-04 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | pH Dependence of Catalysis by Pseudomonas aeruginosa Isochorismate-Pyruvate Lyase: Implications for Transition State Stabilization and the Role of Lysine 42.
Biochemistry, 50, 2011
|
|
3E83
 
 | | Crystal Structure of the the open NaK channel pore | | Descriptor: | CESIUM ION, Potassium channel protein, SODIUM ION | | Authors: | Jiang, Y, Alam, A. | | Deposit date: | 2008-08-19 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of ion selectivity in the NaK channel
Nat.Struct.Mol.Biol., 16, 2009
|
|
3RA7
 
 | |
3RET
 
 | | Salicylate and Pyruvate Bound Structure of the Isochorismate-Pyruvate Lyase K42E Mutant from Pseudomonas aerugionsa | | Descriptor: | 2-HYDROXYBENZOIC ACID, PYRUVIC ACID, Salicylate biosynthesis protein pchB | | Authors: | Olucha, J, Ouellette, A.N, Luo, Q, Lamb, A.L. | | Deposit date: | 2011-04-05 | | Release date: | 2011-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | pH Dependence of Catalysis by Pseudomonas aeruginosa Isochorismate-Pyruvate Lyase: Implications for Transition State Stabilization and the Role of Lysine 42.
Biochemistry, 50, 2011
|
|
3SQX
 
 | |
6AR5
 
 | |
2ZD2
 
 | |
6AZW
 
 | | IDO1/FXB-001116 crystal structure | | Descriptor: | (2R)-N-(4-cyanophenyl)-2-[cis-4-(quinolin-4-yl)cyclohexyl]propanamide, Indoleamine 2,3-dioxygenase 1 | | Authors: | Lewis, H.A, Lammens, A, Steinbacher, S. | | Deposit date: | 2017-09-13 | | Release date: | 2018-03-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Immune-modulating enzyme indoleamine 2,3-dioxygenase is effectively inhibited by targeting its apo-form.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CUL
 
 | | PvdF of pyoverdin biosynthesis is a structurally unique N10-formyltetrahydrofolate-dependent formyltransferase | | Descriptor: | CITRIC ACID, N-(4-{[(2-amino-4-oxo-1,4-dihydroquinazolin-6-yl)methyl]amino}benzene-1-carbonyl)-D-glutamic acid, Pyoverdine synthetase F | | Authors: | Kenjic, N, Hoag, M.R, Moraski, G.C, Caperelli, C.A, Moran, G.R, Lamb, A.L. | | Deposit date: | 2018-03-26 | | Release date: | 2019-02-06 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | PvdF of pyoverdin biosynthesis is a structurally unique N10-formyltetrahydrofolate-dependent formyltransferase.
Arch. Biochem. Biophys., 664, 2019
|
|
8PJA
 
 | | FKBP51FK1 F67E/K58 (i, i+9) in complex with SAFit1 | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Charalampidou, A, Hausch, F. | | Deposit date: | 2023-06-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Automated Flow Peptide Synthesis Enables Engineering of Proteins with Stabilized Transient Binding Pockets.
Acs Cent.Sci., 10, 2024
|
|
6AR3
 
 | | Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications (RT/Duplex (Se-Met)) | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA, GsI-IIC RT, ... | | Authors: | Stamos, J.L, Lentzsch, A.M, Lambowitz, A.M. | | Deposit date: | 2017-08-21 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications.
Mol. Cell, 68, 2017
|
|
8PJ8
 
 | | FKBP51FK1 F67E/K60Orn (i, i+7) in complex with SAFit1 | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Charalampidou, A, Hausch, F. | | Deposit date: | 2023-06-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Automated Flow Peptide Synthesis Enables Engineering of Proteins with Stabilized Transient Binding Pockets.
Acs Cent.Sci., 10, 2024
|
|
6AR1
 
 | | Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications (RT/Duplex (Nat)) | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA, GsI-IIC RT, ... | | Authors: | Stamos, J.L, Lentzsch, A.M, Lambowitz, A.M. | | Deposit date: | 2017-08-21 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structure of a Thermostable Group II Intron Reverse Transcriptase with Template-Primer and Its Functional and Evolutionary Implications.
Mol. Cell, 68, 2017
|
|
6C4R
 
 | | Staphylopine dehydrogenase (SaODH) - Apo | | Descriptor: | GLYCEROL, SULFATE ION, Staphylopine dehydrogenase | | Authors: | McFarlane, J.S, Davis, C.L, Lamb, A.L. | | Deposit date: | 2018-01-12 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Staphylopine, pseudopaline, and yersinopine dehydrogenases: A structural and kinetic analysis of a new functional class of opine dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
6C4N
 
 | | Pseudopaline dehydrogenase (PaODH) - NADP+ bound | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pseudopaline dehydrogenase | | Authors: | McFarlane, J.S, Davis, C.L, Lamb, A.L. | | Deposit date: | 2018-01-12 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Staphylopine, pseudopaline, and yersinopine dehydrogenases: A structural and kinetic analysis of a new functional class of opine dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
6DQO
 
 | |
6C4T
 
 | | Staphylopine dehydrogenase (SaODH) - NADP+ bound | | Descriptor: | GLYCEROL, Staphylopine dehydrogenase, [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4S)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | McFarlane, J.S, Davis, C.L, Lamb, A.L. | | Deposit date: | 2018-01-12 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Staphylopine, pseudopaline, and yersinopine dehydrogenases: A structural and kinetic analysis of a new functional class of opine dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
6C4M
 
 | | Yersinopine dehydrogenase (YpODH) - NADP+ bound | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Yersinopine dehydrogenase | | Authors: | McFarlane, J.S, Davis, C.L, Lamb, A.L. | | Deposit date: | 2018-01-12 | | Release date: | 2018-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Staphylopine, pseudopaline, and yersinopine dehydrogenases: A structural and kinetic analysis of a new functional class of opine dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
6C4L
 
 | | Yersinopine dehydrogenase (YpODH) - Apo | | Descriptor: | Yersinopine dehydrogenase | | Authors: | McFarlane, J.S, Davis, C.L, Lamb, A.L. | | Deposit date: | 2018-01-12 | | Release date: | 2018-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Staphylopine, pseudopaline, and yersinopine dehydrogenases: A structural and kinetic analysis of a new functional class of opine dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
1IIG
 
 | | STRUCTURE OF TRYPANOSOMA BRUCEI BRUCEI TRIOSEPHOSPHATE ISOMERASE COMPLEXED WITH 3-PHOSPHONOPROPIONATE | | Descriptor: | 3-PHOSPHONOPROPANOIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Noble, M.E, Wierenga, R.K, Lambeir, A.M, Opperdoes, F.R, Thunnissen, A.M, Kalk, K.H, Groendijk, H, Hol, W.G.J. | | Deposit date: | 2001-04-23 | | Release date: | 2001-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The adaptability of the active site of trypanosomal triosephosphate isomerase as observed in the crystal structures of three different complexes.
Proteins, 10, 1991
|
|
