5O1O
 
 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain with proline bound. | | Descriptor: | 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, mitochondrial, ... | | Authors: | Kopec, J, Rembeza, E, Pena, I.A, Mathea, S, Velupillai, S, Strain-Damerell, C, Goubin, S, Kupinska, K, Talon, R, Collins, P, Krojer, T, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, von Delft, F, Arruda, P, Yue, W.W. | | Deposit date: | 2017-05-18 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain with proline bound.
To Be Published
|
|
6H50
 
 | | Crystal structure of human KDM5B in complex with compound 34a | | Descriptor: | 1,2-ETHANEDIOL, 8-[4-(1-methylpiperidin-4-yl)pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, Velupillai, S, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
6H51
 
 | | Crystal structure of human KDM5B in complex with compound 34f | | Descriptor: | 1,2-ETHANEDIOL, 8-[4-[1-(cyclobutylmethyl)piperidin-4-yl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, Velupillai, S, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
5V6C
 
 | | Crystal Structure of the Second beta-Prism Domain of RbmC from V. cholerae | | Descriptor: | GLYCEROL, Hemolysin-related protein | | Authors: | De, S, Kaus, K, Sinclair, S, Case, B.C, Olson, R. | | Deposit date: | 2017-03-16 | | Release date: | 2018-01-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of mammalian glycan targeting by Vibrio cholerae cytolysin and biofilm proteins.
PLoS Pathog., 14, 2018
|
|
6FM5
 
 | | Crystal structure of self-complemented CsuA/B major subunit from archaic chaperone-usher Csu pili of Acinetobacter baumannii | | Descriptor: | CsuA/B,CsuA/B,CsuA/B,CsuA/B | | Authors: | Pakharukova, N.A, Tuitilla, M, Paavilainen, S, Zavialov, A.V. | | Deposit date: | 2018-01-30 | | Release date: | 2018-09-26 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Archaic and alternative chaperones preserve pilin folding energy by providing incomplete structural information.
J. Biol. Chem., 293, 2018
|
|
6FQ0
 
 | | Crystal structure of the CsuC-CsuA/B chaperone-subunit preassembly complex of the archaic chaperone-usher Csu pili of Acinetobacter baumannii | | Descriptor: | CsuA/B,CsuA/B, CsuC | | Authors: | Pakharukova, N.A, Tuitilla, M, Paavilainen, S, Zavialov, A.V. | | Deposit date: | 2018-02-12 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Archaic and alternative chaperones preserve pilin folding energy by providing incomplete structural information.
J. Biol. Chem., 293, 2018
|
|
6FQA
 
 | | Crystal structure of the CsuC-CsuA/B chaperone-subunit preassembly complex of the archaic chaperone-usher Csu pili of Acinetobacter baumannii | | Descriptor: | CsuA/B,CsuA/B, CsuC | | Authors: | Parilova, O, Pakharukova, N.A, Malmi, H, Tuitilla, M, Paavilainen, S, Zavialov, A.V. | | Deposit date: | 2018-02-13 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Archaic and alternative chaperones preserve pilin folding energy by providing incomplete structural information.
J. Biol. Chem., 293, 2018
|
|
6H4Z
 
 | | Crystal structure of human KDM5B in complex with compound 16a | | Descriptor: | 1,2-ETHANEDIOL, 8-[4-[2-[4-(3-chlorophenyl)piperidin-1-yl]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, Velupillai, S, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
5V6F
 
 | | Crystal Structure of the Second beta-Prism Domain of RbmC from V. cholerae bound to Mannotriose | | Descriptor: | Hemolysin-related protein, SULFATE ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose | | Authors: | De, S, Kaus, K, Sinclair, S, Case, B.C, Olson, R. | | Deposit date: | 2017-03-16 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of mammalian glycan targeting by Vibrio cholerae cytolysin and biofilm proteins.
PLoS Pathog., 14, 2018
|
|
6H52
 
 | | Crystal structure of human KDM5B in complex with compound 34g | | Descriptor: | 1,2-ETHANEDIOL, 8-[4-(1-cyclopentylpiperidin-4-yl)pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, Velupillai, S, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
5FVJ
 
 | | Crystal structure of TacT (tRNA acetylating toxin) from Salmonella | | Descriptor: | ACETYL COENZYME *A, PUTATIVE ACETYLTRANSFERASE | | Authors: | Przydacz, M, Wong, C.T, Cheverton, A.M, Gollan, B, Mylona, A, Helaine, S, Hare, S.A. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Salmonella Toxin Promotes Persister Formation Through Acetylation of tRNA.
Mol.Cell, 63, 2016
|
|
5MP0
 
 | | Human m7GpppN-mRNA Hydrolase (DCP2, NUDT20) Catalytic Domain | | Descriptor: | 1,2-ETHANEDIOL, m7GpppN-mRNA hydrolase | | Authors: | Mathea, S, Salah, E, Velupillai, S, Tallant, C, Pike, A.C.W, Bushell, S.R, Faust, B, Wang, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Huber, K. | | Deposit date: | 2016-12-15 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Human m7GpppN-mRNA Hydrolase (DCP2, NUDT20) Catalytic Domain
To Be Published
|
|
2M33
 
 | | Solution NMR structure of full-length oxidized microsomal rabbit cytochrome b5 | | Descriptor: | Cytochrome b5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Subramanian, V, Ahuja, S, Popovych, N, Huang, R, Le Clair, S.V, Jahr, N, Soong, R, Xu, J, Yamamoto, K, Nanga, R.P, Im, S, Waskell, L, Ramamoorthy, A. | | Deposit date: | 2013-01-08 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of full-length mammalian cytochrome b5
To be Published
|
|
6G96
 
 | | Crystal structure of TacT3 (tRNA acetylating toxin) from Salmonella | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase, BICINE, ... | | Authors: | Grabe, G.J, Rycroft, J.A, Gollan, B, Hall, A, Cheverton, A.M, Larrouy-Maumus, G, Hare, S.A, Helaine, S. | | Deposit date: | 2018-04-10 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4766078 Å) | | Cite: | Activity of acetyltransferase toxins involved in Salmonella persister formation during macrophage infection.
Nat Commun, 9, 2018
|
|
6GK0
 
 | | HUMAN DIHYDROOROTATE DEHYDROGENASE IN COMPLEX WITH CLASS III HISTONE DEACETYLASE INHIBITOR | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, 4-~{tert}-butyl-~{N}-[[4-[5-(dimethylamino)pentanoylamino]phenyl]carbamothioyl]benzamide, ACETIC ACID, ... | | Authors: | Hakansson, M, Ladds, M.J.G.W, Walse, B, Lain, S. | | Deposit date: | 2018-05-17 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Exploitation of dihydroorotate dehydrogenase (DHODH) and p53 activation as therapeutic targets: A case study in polypharmacology.
J.Biol.Chem., 295, 2020
|
|
5D6H
 
 | | Crystal structure of CsuC-CsuA/B chaperone-major subunit pre-assembly complex from Csu biofilm-mediating pili of Acinetobacter baumannii | | Descriptor: | CsuA/B, CsuC | | Authors: | Pakharukova, N.A, Tuitilla, M, Paavilainen, S, Zavialov, A. | | Deposit date: | 2015-08-12 | | Release date: | 2015-11-04 | | Last modified: | 2015-12-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight into Archaic and Alternative Chaperone-Usher Pathways Reveals a Novel Mechanism of Pilus Biogenesis.
Plos Pathog., 11, 2015
|
|
5O1N
 
 | | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain with N-[(2S)-2-Pyrrolidinylmethyl]-trifluoromethanesulfonamide bound | | Descriptor: | 1,1,1-tris(fluoranyl)-~{N}-[[(2~{S})-pyrrolidin-2-yl]methyl]methanesulfonamide, 1,2-ETHANEDIOL, Alpha-aminoadipic semialdehyde synthase, ... | | Authors: | Kopec, J, Rembeza, E, Pena, I.A, Strain-Damerell, C, Goubin, S, Sethi, R, Velupillai, S, Talon, R, Collins, P, Krojer, T, McLaughlin, M, Burgess-Brown, N, Arrowsmith, C, Edwards, A, Bountra, C, Brennan, P, von Delft, F, Arruda, P, Yue, W.W. | | Deposit date: | 2017-05-18 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of human aminoadipate semialdehyde synthase, saccharopine dehydrogenase domain with N-[(2S)-2-Pyrrolidinylmethyl]-trifluoromethanesulfonamide bound
To Be Published
|
|
2WMW
 
 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1-[(2S)-4-(5-BROMO-1H-PYRAZOLO[3,4-B]PYRIDIN-4-YL)MORPHOLIN-2-YL]METHANAMINE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, Klair, S, Burns, S, Boxall, K, Cherry, M, Fisher, M, Westwood, I.M, Walton, M.I, McHardy, T, Cheung, K.-M.J, Van Montfort, R, Williams, D, Aherne, G.W, Garrett, M.D, Reader, J, Collins, I. | | Deposit date: | 2009-07-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Identification of Inhibitors of Checkpoint Kinase 1 Through Template Screening.
J.Med.Chem., 52, 2009
|
|
2WMV
 
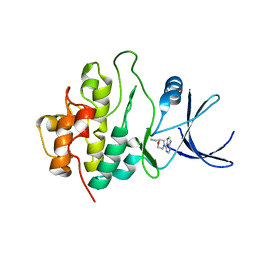 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1-[(2S)-4-(7H-PURIN-6-YL)MORPHOLIN-2-YL]METHANAMINE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, Klair, S, Burns, S, Boxall, K, Cherry, M, Fisher, M, Westwood, I.M, Walton, M.I, McHardy, T, Cheung, K.-M.J, Van Montfort, R, Williams, D, Aherne, G.W, Garrett, M.D, Reader, J, Collins, I. | | Deposit date: | 2009-07-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Identification of Inhibitors of Checkpoint Kinase 1 Through Template Screening.
J.Med.Chem., 52, 2009
|
|
2WMT
 
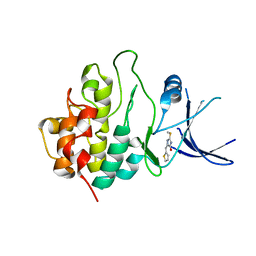 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 2-(methylsulfanyl)-5-(thiophen-2-ylmethyl)-1H-imidazol-4-ol, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, Klair, S, Burns, S, Boxall, K, Cherry, M, Fisher, M, Westwood, I.M, Walton, M.I, McHardy, T, Cheung, K.-M.J, Van Montfort, R, Williams, D, Aherne, G.W, Garrett, M.D, Reader, J, Collins, I. | | Deposit date: | 2009-07-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification of Inhibitors of Checkpoint Kinase 1 Through Template Screening.
J.Med.Chem., 52, 2009
|
|
2WMX
 
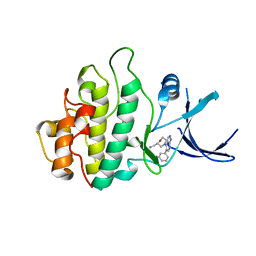 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1-[(2S)-4-(5-phenyl-1H-pyrazolo[3,4-b]pyridin-4-yl)morpholin-2-yl]methanamine, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, Klair, S, Burns, S, Boxall, K, Cherry, M, Fisher, M, Westwood, I.M, Walton, M.I, McHardy, T, Cheung, K.-M.J, Van Montfort, R, Williams, D, Aherne, G.W, Garrett, M.D, Reader, J, Collins, I. | | Deposit date: | 2009-07-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification of Inhibitors of Checkpoint Kinase 1 Through Template Screening.
J.Med.Chem., 52, 2009
|
|
2WMU
 
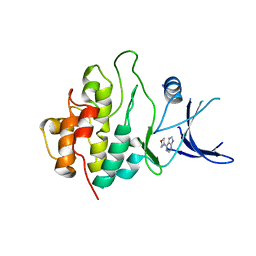 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 6-MORPHOLIN-4-YL-9H-PURINE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, Klair, S, Burns, S, Boxall, K, Cherry, M, Fisher, M, Westwood, I.M, Walton, M.I, McHardy, T, Cheung, K.-M.J, Van Montfort, R, Williams, D, Aherne, G.W, Garrett, M.D, Reader, J, Collins, I. | | Deposit date: | 2009-07-03 | | Release date: | 2009-07-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of Inhibitors of Checkpoint Kinase 1 Through Template Screening.
J.Med.Chem., 52, 2009
|
|
6ET4
 
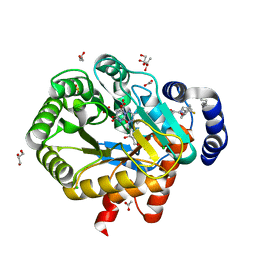 | | HUMAN DIHYDROOROTATE DEHYDROGENASE IN COMPLEX WITH NOVEL INHIBITOR | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Hakansson, M, Walse, B, Gustavsson, A.-L, Lain, S. | | Deposit date: | 2017-10-25 | | Release date: | 2018-03-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A DHODH inhibitor increases p53 synthesis and enhances tumor cell killing by p53 degradation blockage.
Nat Commun, 9, 2018
|
|
6FJY
 
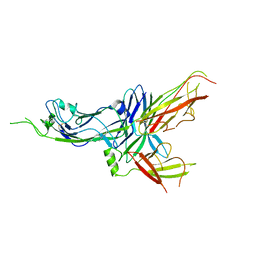 | | Crystal structure of CsuC-CsuE chaperone-tip adhesion subunit pre-assembly complex from archaic chaperone-usher Csu pili of Acinetobacter baumannii | | Descriptor: | CsuC, Protein CsuE | | Authors: | Pakharukova, N.A, Tuitilla, M, Paavilainen, S, Zavialov, A.V. | | Deposit date: | 2018-01-23 | | Release date: | 2018-05-16 | | Last modified: | 2018-09-19 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis forAcinetobacter baumanniibiofilm formation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5NLB
 
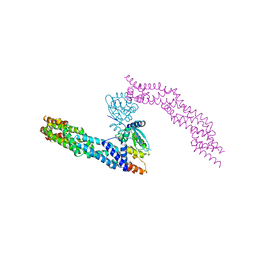 | | Crystal structure of human CUL3 N-terminal domain bound to KEAP1 BTB and 3-box | | Descriptor: | Cullin-3, Kelch-like ECH-associated protein 1 | | Authors: | Adamson, R, Krojer, T, Pinkas, D.M, Bartual, S.G, Burgess-Brown, N.A, Borkowska, O, Chalk, R, Newman, J.A, Kopec, J, Dixon-Clarke, S.E, Mathea, S, Sethi, R, Velupillai, S, Mackinnon, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2017-04-04 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural and biochemical characterization establishes a detailed understanding of KEAP1-CUL3 complex assembly.
Free Radic Biol Med, 204, 2023
|
|
