8XYW
 
 | | De novo designed protein Trx-3 | | Descriptor: | De novo designed Trx-3 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8XYR
 
 | | De novo designed protein GPX4-2 | | Descriptor: | De novo designed GPX4-2 | | Authors: | Liu, L.J, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8XYV
 
 | | De novo designed protein 0705-5 | | Descriptor: | De novo designed protein 0705-5 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8XYU
 
 | | De novo designed protein GPX4-3 | | Descriptor: | De novo designed GPX4-3 | | Authors: | Guo, Z, Liu, J.L, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8XYS
 
 | | De novo designed protein GPX4-1 | | Descriptor: | De novo designed GPX4-1 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
8XYT
 
 | | De novo designed protein GPX4-4 | | Descriptor: | De novo designed GPX4-4 | | Authors: | Liu, J.L, Guo, Z, Lai, L.H. | | Deposit date: | 2024-01-20 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | All-Atom Protein Sequence Design Based on Geometric Deep Learning.
Angew.Chem.Int.Ed.Engl., 2024
|
|
7C0N
 
 | | Crystal structure of a self-assembling galactosylated peptide homodimer | | Descriptor: | SULFATE ION, Self-assembling galactosylated tyrosine-rich peptide, beta-D-galactopyranose | | Authors: | He, C, Wu, S, Chi, C, Zhang, W, Ma, M, Lai, L, Dong, S. | | Deposit date: | 2020-05-01 | | Release date: | 2020-10-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Glycopeptide Self-Assembly Modulated by Glycan Stereochemistry through Glycan-Aromatic Interactions.
J.Am.Chem.Soc., 142, 2020
|
|
4A5V
 
 | | Solution structure ensemble of the two N-terminal apple domains (residues 58-231) of Toxoplasma gondii microneme protein 4 | | Descriptor: | MICRONEMAL PROTEIN 4 | | Authors: | Marchant, J, Cowper, B, Liu, Y, Lai, L, Pinzan, C, Marq, J.B, Friedrich, N, Sawmynaden, K, Chai, W, Childs, R.A, Saouros, S, Simpson, P, Barreira, M.C.R, Feizi, T, Soldati-Favre, D, Matthews, S. | | Deposit date: | 2011-10-28 | | Release date: | 2012-04-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Galactose Recognition by the Apicomplexan Parasite Toxoplasma Gondii.
J.Biol.Chem., 287, 2012
|
|
3TKK
 
 | | Crystal Structure Analysis of a recombinant predicted acetamidase/ formamidase from the thermophile thermoanaerobacter tengcongensis | | Descriptor: | CALCIUM ION, Predicted acetamidase/formamidase, ZINC ION | | Authors: | Qian, M, Huang, Q, Wu, G, Lai, L, Tang, Y, Pei, J, Kusunoki, M. | | Deposit date: | 2011-08-26 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure Analysis of a Recombinant Predicted Acetamidase/Formamidase from the Thermophile Thermoanaerobacter tengcongensis.
PROTEIN J., 31, 2012
|
|
2KI0
 
 | | NMR Structure of a de novo designed beta alpha beta | | Descriptor: | DS119 | | Authors: | Liang, H, Chen, H, Fan, K, Wei, P, Guo, X, Jin, C, Zeng, C, Tang, C, Lai, L. | | Deposit date: | 2009-04-18 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | De novo design of a beta alpha beta motif.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3OX8
 
 | | Crystal Structure of HLA A*02:03 Bound to HBV Core 18-27 | | Descriptor: | 10mer peptide from Pre-core-protein, Beta-2-microglobulin, MHC class I antigen | | Authors: | Liu, J, Chen, Y, Lai, L, Ren, E. | | Deposit date: | 2010-09-21 | | Release date: | 2011-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural insights into the binding of hepatitis B virus core peptide to HLA-A2 alleles: Towards designing better vaccines.
Eur.J.Immunol., 41, 2011
|
|
3OXR
 
 | | Crystal Structure of HLA A*02:06 Bound to HBV Core 18-27 | | Descriptor: | 10mer peptide from Pre-core-protein, Beta-2-microglobulin, MHC class I antigen | | Authors: | Liu, J, Chen, Y, Lai, L, Ren, E. | | Deposit date: | 2010-09-21 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the binding of hepatitis B virus core peptide to HLA-A2 alleles: Towards designing better vaccines.
Eur.J.Immunol., 41, 2011
|
|
3OXS
 
 | | Crystal Structure of HLA A*02:07 Bound to HBV Core 18-27 | | Descriptor: | 10mer peptide from Pre-core-protein, Beta-2-microglobulin, MHC class I antigen | | Authors: | Liu, J, Chen, Y, Lai, L, Ren, E. | | Deposit date: | 2010-09-22 | | Release date: | 2011-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the binding of hepatitis B virus core peptide to HLA-A2 alleles: Towards designing better vaccines.
Eur.J.Immunol., 41, 2011
|
|
5HQD
 
 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Roesser, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | Deposit date: | 2016-01-21 | | Release date: | 2016-02-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
5F81
 
 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Roessler, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | Deposit date: | 2015-12-08 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
5HL4
 
 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | Descriptor: | COBALT HEXAMMINE(III), FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Roessler, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
6DNA
 
 | | Crystal structure of T110A mutant human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
6XNN
 
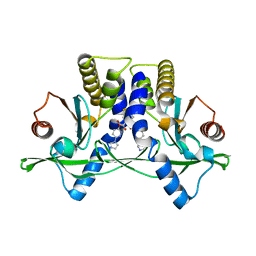 | | Crystal Structure of Mouse STING CTD complex with SR-717. | | Descriptor: | 4,5-difluoro-2-{[6-(1H-imidazol-1-yl)pyridazine-3-carbonyl]amino}benzoic acid, Stimulator of interferon genes protein | | Authors: | Chin, E.N, Yu, C, Wolan, D.W, Petrassi, H.M, Lairson, L.L. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Antitumor activity of a systemic STING-activating non-nucleotide cGAMP mimetic.
Science, 369, 2020
|
|
6XNP
 
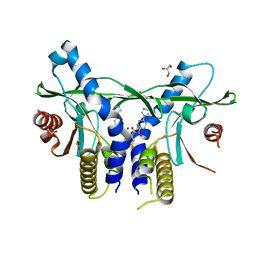 | | Crystal Structure of Human STING CTD complex with SR-717 | | Descriptor: | 1,2-ETHANEDIOL, 4,5-difluoro-2-{[6-(1H-imidazol-1-yl)pyridazine-3-carbonyl]amino}benzoic acid, GLYCEROL, ... | | Authors: | Chin, E.N, Yu, C, Wolan, D.W, Petrassi, H.M, Lairson, L.L. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Antitumor activity of a systemic STING-activating non-nucleotide cGAMP mimetic.
Science, 369, 2020
|
|
6DND
 
 | | Crystal structure of wild-type (WT) human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
6DNB
 
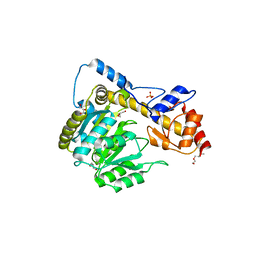 | | Crystal structure of T110A:S256A mutant human Glutamate oxaloacetate transaminase 1 (GOT1) | | Descriptor: | Aspartate aminotransferase, cytoplasmic, GLYCEROL, ... | | Authors: | Assar, Z, Holt, M.C, Stein, A.J, Lairson, L, Lyssiotis, C.A. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical Characterization and Structure-Based Mutational Analysis Provide Insight into the Binding and Mechanism of Action of Novel Aspartate Aminotransferase Inhibitors.
Biochemistry, 57, 2018
|
|
6TKV
 
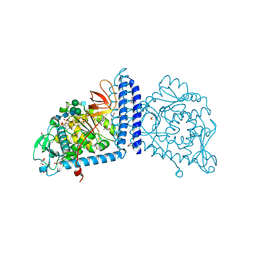 | | Crystal structure of the human FUT8 in complex with GDP and a biantennary complex N-glycan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-(1,6)-fucosyltransferase, GLYCEROL, ... | | Authors: | Garcia-Garcia, A, Ceballos-Laita, L, Serna, L, Artschwager, R, Reichardt, N.C, Corzana, F, Hurtado-Guerrero, R. | | Deposit date: | 2019-11-29 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for substrate specificity and catalysis of alpha 1,6-fucosyltransferase.
Nat Commun, 11, 2020
|
|
6S22
 
 | | Crystal structure of the TgGalNAc-T3 in complex with UDP, manganese and FGF23c | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | de las Rivas, M, Daniel, E.J.P, Narimatsu, Y, Companon, I, Kato, K, Hermosilla, P, Thureau, A, Ceballos-Laita, L, Coelho, H, Bernado, P, Marcelo, F, Hansen, L, Lostao, A, Corzana, F, Clausen, H, Gerken, T.A, Hurtado-Guerrero, R. | | Deposit date: | 2019-06-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Molecular basis for fibroblast growth factor 23 O-glycosylation by GalNAc-T3.
Nat.Chem.Biol., 16, 2020
|
|
6S24
 
 | | Crystal structure of the TgGalNAc-T3 in complex with UDP, manganese and the peptide 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ALA-THR-GLY-ALA-GLY-ALA-GLY-ALA-GLY-THR-THR-PRO-GLY-PRO, ... | | Authors: | de las Rivas, M, Daniel, E.J.P, Narimatsu, Y, Companon, I, Kato, K, Hermosilla, P, Thureau, A, Ceballos-Laita, L, Coelho, H, Bernado, P, Marcelo, F, Hansen, L, Lostao, A, Corzana, F, Clausen, H, Gerken, T.A, Hurtado-Guerrero, R. | | Deposit date: | 2019-06-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Molecular basis for fibroblast growth factor 23 O-glycosylation by GalNAc-T3.
Nat.Chem.Biol., 16, 2020
|
|
2WQQ
 
 | | Crystallographic analysis of monomeric CstII | | Descriptor: | ALPHA-2,3-/2,8-SIALYLTRANSFERASE, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, DI(HYDROXYETHYL)ETHER | | Authors: | Chan, P.H.W, Lairson, L.L, Lee, H.J, Wakarchuk, W.W, Strynadka, N.C.J, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2009-08-25 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | NMR Spectroscopic Characterization of the Sialyltransferase Cstii from Camplyobacter Jejuni: Histidine 188 is the General Base.
Biochemistry, 48, 2009
|
|
