8VDW
 
 | |
8VCW
 
 | | X-Ray Crystal Structure of the biotin synthase from B. obeum | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, Biotin synthase, ... | | Authors: | Lachowicz, J.C, Grove, T.L. | | Deposit date: | 2023-12-14 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery of a Biotin Synthase That Utilizes an Auxiliary 4Fe-5S Cluster for Sulfur Insertion.
J.Am.Chem.Soc., 146, 2024
|
|
9DFU
 
 | |
9DFW
 
 | |
9DFN
 
 | |
9DGW
 
 | |
6CCJ
 
 | |
6CE5
 
 | |
6CUS
 
 | |
6CW4
 
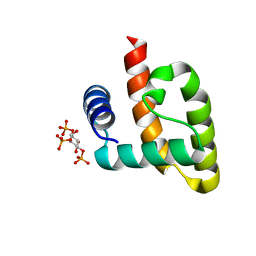 | | HADDOCK structure of the Rous sarcoma virus matrix protein (M-domain) in complex with inositol 1,3,5-trisphosphate | | Descriptor: | (1R,2S,3r,4R,5S,6s)-2,4,6-trihydroxycyclohexane-1,3,5-triyl tris[dihydrogen (phosphate)], Matrix protein p19 | | Authors: | Vlach, J, Saad, J.S. | | Deposit date: | 2018-03-29 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for targeting avian sarcoma virus Gag polyprotein to the plasma membrane for virus assembly.
J. Biol. Chem., 293, 2018
|
|
6CV8
 
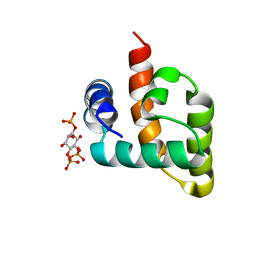 | | HADDOCK structure of the Rous sarcoma virus matrix protein (M-domain) in complex with inositol 1,4,5-trisphosphate | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Matrix protein p19 | | Authors: | Vlach, J, Saad, J.S. | | Deposit date: | 2018-03-27 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for targeting avian sarcoma virus Gag polyprotein to the plasma membrane for virus assembly.
J. Biol. Chem., 293, 2018
|
|
2F77
 
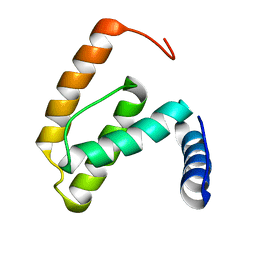 | | Solution structure of the R55F mutant of M-PMV matrix protein (p10) | | Descriptor: | Core protein p10 | | Authors: | Vlach, J, Lipov, J, Veverka, V, Lang, J, Srb, P, Rumlova, M, Hunter, E, Ruml, T, Hrabal, R. | | Deposit date: | 2005-11-30 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | D-retrovirus morphogenetic switch driven by the targeting signal accessibility to Tctex-1 of dynein.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2F76
 
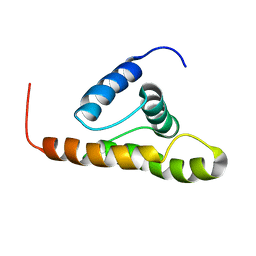 | | Solution structure of the M-PMV wild type matrix protein (p10) | | Descriptor: | Core protein p10 | | Authors: | Vlach, J, Lipov, J, Veverka, V, Lang, J, Srb, P, Rumlova, M, Hunter, E, Ruml, T, Hrabal, R. | | Deposit date: | 2005-11-30 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | D-retrovirus morphogenetic switch driven by the targeting signal accessibility to Tctex-1 of dynein.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2LYA
 
 | |
2MGU
 
 | |
2LYB
 
 | |
4NCW
 
 | | foldon domain wild type C-conjugate | | Descriptor: | Fibritin, N,N',N''-triethylbenzene-1,3,5-tricarboxamide | | Authors: | Graewert, M.A, Berthelmann, A, Lach, J, Groll, M, Eichler, J. | | Deposit date: | 2013-10-25 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Versatile C(3)-symmetric scaffolds and their use for covalent stabilization of the foldon trimer.
Org.Biomol.Chem., 12, 2014
|
|
4NCV
 
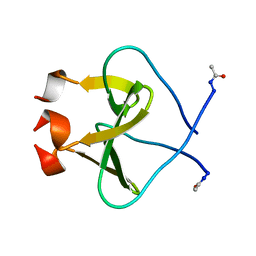 | | Foldon domain wild type N-conjugate | | Descriptor: | Fibritin | | Authors: | Graewert, M.A, Berthelmann, A, Lach, J, Groll, M, Eichler, J. | | Deposit date: | 2013-10-25 | | Release date: | 2014-03-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Versatile C(3)-symmetric scaffolds and their use for covalent stabilization of the foldon trimer.
Org.Biomol.Chem., 12, 2014
|
|
3TTI
 
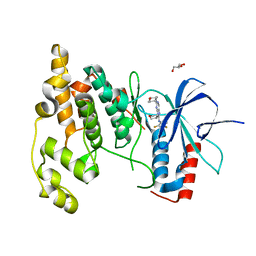 | | Crystal Structure of JNK3 complexed with CC-930, an orally active anti-fibrotic JNK inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 10, trans-4-({9-[(3S)-tetrahydrofuran-3-yl]-8-[(2,4,6-trifluorophenyl)amino]-9H-purin-2-yl}amino)cyclohexanol | | Authors: | Plantevin-Krenitsky, V, Nadolny, L, Delgado, M, Ayala, L, Clareen, S, Hilgraf, R, Albers, R, Hegde, S, D'Sidocky, N, Sapienza, J, Wright, J, McCarrick, M, Bahmanyar, S, Chamberlain, P, Delker, S.L, Muir, J, Giegel, D, Xu, L, Celeridad, M, Lachowitzer, J, Bennett, B, Moghaddam, M, Khatsenko, O, Katz, J, Fan, R, Bai, A, Tang, Y, Shirley, M.A, Benish, B, Bodine, T, Blease, K, Raymon, H, Cathers, B.E, Satoh, Y. | | Deposit date: | 2011-09-14 | | Release date: | 2012-02-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of CC-930, an orally active anti-fibrotic JNK inhibitor.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
7N7I
 
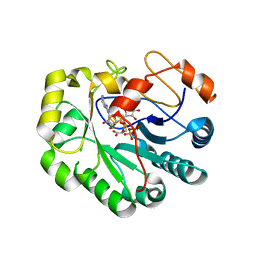 | | X-ray crystal structure of Viperin-like enzyme from Trichoderma virens | | Descriptor: | IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Grove, T.L, Almo, S.C, Bonanno, J.B, Lachowicz, J.C, Gizzi, A.G. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural Insight into the Substrate Scope of Viperin and Viperin-like Enzymes from Three Domains of Life.
Biochemistry, 60, 2021
|
|
8VPO
 
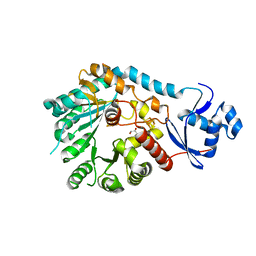 | | X-Ray Crystal Structure of TigE from Paramaledivibacter caminithermalis | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, Radical SAM core domain-containing protein | | Authors: | Grove, T.L, Lachowicz, J.C, Zizola, C. | | Deposit date: | 2024-01-16 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural, Biochemical, and Bioinformatic Basis for Identifying Radical SAM Cyclopropyl Synthases.
Acs Chem.Biol., 19, 2024
|
|
8DKU
 
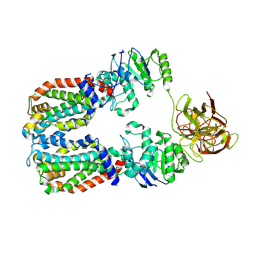 | | CryoEM structure of the A. aeolicus WzmWzt transporter bound to the native O antigen | | Descriptor: | 6-deoxy-3-O-methyl-alpha-D-mannopyranose-(1-3)-beta-D-rhamnopyranose-(1-2)-alpha-D-rhamnopyranose, ABC transporter, Transport permease protein | | Authors: | Spellmon, N, Muszynski, A, Vlach, J, Zimmer, J. | | Deposit date: | 2022-07-06 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis for polysaccharide recognition and modulated ATP hydrolysis by the O antigen ABC transporter.
Nat Commun, 13, 2022
|
|
8DNC
 
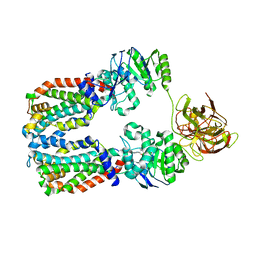 | | CryoEM structure of the A. aeolicus WzmWzt transporter bound to the native O antigen and ADP | | Descriptor: | 6-deoxy-3-O-methyl-alpha-D-mannopyranose-(1-3)-beta-D-rhamnopyranose-(1-2)-alpha-D-rhamnopyranose, ABC transporter, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Spellmon, N, Muszynski, A, Vlach, J, Zimmer, J. | | Deposit date: | 2022-07-11 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for polysaccharide recognition and modulated ATP hydrolysis by the O antigen ABC transporter.
Nat Commun, 13, 2022
|
|
7N7H
 
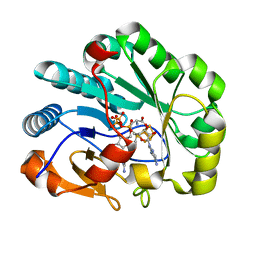 | | X-ray crystal structure of Viperin-like enzyme from Nematostella vectensis | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE, ... | | Authors: | Grove, T.L, Almo, S.C, Bonanno, J.B, Lachowicz, J.C, Gizzi, A.G. | | Deposit date: | 2021-06-10 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural Insight into the Substrate Scope of Viperin and Viperin-like Enzymes from Three Domains of Life.
Biochemistry, 60, 2021
|
|
2XMY
 
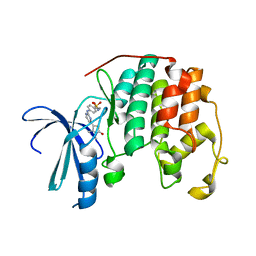 | | Discovery and Characterisation of 2-Anilino-4-(thiazol-5-yl) pyrimidine Transcriptional CDK Inhibitors as Anticancer Agents | | Descriptor: | 4-[4-(3,4-DIMETHYL-2-OXO-2,3-DIHYDRO-THIAZOL-5-YL)-PYRIMIDIN-2-YLAMINO]-N-(2-METHOXY-ETHYL)-BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Wang, S, Griffiths, G, Midgley, C.A, Barnett, A.L, Cooper, M, Grabarek, J, Ingram, L, Jackson, W, Kontopidis, G, McClue, S.J, McInnes, C, McLachlan, J, Meades, C, Mezna, M, Stuart, I, Thomas, M.P, Zheleva, D.I, Lane, D.P, Jackson, R.C, Glover, D.M, Blake, D.G, Fischer, P.M. | | Deposit date: | 2010-07-29 | | Release date: | 2010-11-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Characterisation of 2-Anilino-4-(Thiazol-5-Yl)Pyrimidine Transcriptional Cdk Inhibitors as Anticancer Agents
Chem.Biol., 17, 2010
|
|
