2YJT
 
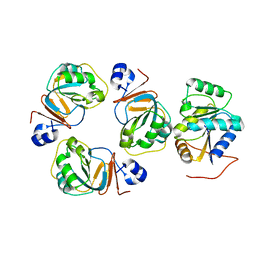 | |
1EK9
 
 | | 2.1A X-RAY STRUCTURE OF TOLC: AN INTEGRAL OUTER MEMBRANE PROTEIN AND EFFLUX PUMP COMPONENT FROM ESCHERICHIA COLI | | Descriptor: | OUTER MEMBRANE PROTEIN TOLC | | Authors: | Koronakis, V, Sharff, A.J, Koronakis, E, Luisi, B, Hughes, C. | | Deposit date: | 2000-03-07 | | Release date: | 2000-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export.
Nature, 405, 2000
|
|
2WVA
 
 | | Structural insights into the pre-reaction state of pyruvate decarboxylase from Zymomonas mobilis | | Descriptor: | 2-{1-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-5-METHYL-1H-1,2,3-TRIAZOL-4-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, MAGNESIUM ION, PYRUVATE DECARBOXYLASE, ... | | Authors: | Pei, X.Y, Erixon, K, Luisi, B.F, Leeper, F.J. | | Deposit date: | 2009-10-15 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights Into the Pre-Reaction State of Pyruvate Decarboxylase from Zymomonas Mobilis
Biochemistry, 49, 2010
|
|
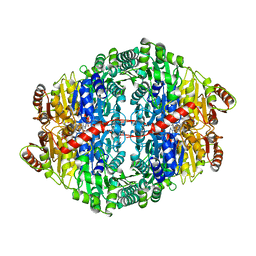
5O5O
 
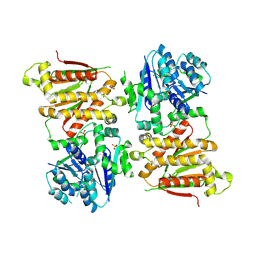 | | X-ray crystal structure of RapZ from Escherichia coli (P32 space group) | | Descriptor: | RNase adapter protein RapZ, SULFATE ION | | Authors: | Gonzalez, G.M, Durica-Mitic, S, Hardwick, S.W, Moncrieffe, M, Resch, M, Neumann, P, Ficner, R, Gorke, B, Luisi, B.F. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-30 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Structural insights into RapZ-mediated regulation of bacterial amino-sugar metabolism.
Nucleic Acids Res., 45, 2017
|
|
5O5Q
 
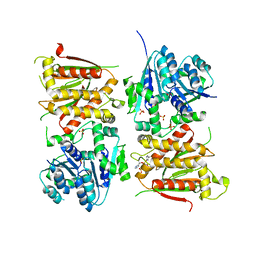 | | X-ray crystal structure of RapZ from Escherichia coli (P3221 space group) | | Descriptor: | RNase adapter protein RapZ, SULFATE ION | | Authors: | Gonzalez, G.M, Durica-Mitic, S, Hardwick, S.W, Moncrieffe, M, Resch, M, Neumann, P, Ficner, R, Gorke, B, Luisi, B.F. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-30 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural insights into RapZ-mediated regulation of bacterial amino-sugar metabolism.
Nucleic Acids Res., 45, 2017
|
|
1CHC
 
 | |
2VMK
 
 | | Crystal Structure of E. coli RNase E Apoprotein - Catalytic Domain | | Descriptor: | RIBONUCLEASE E, SULFATE ION, ZINC ION | | Authors: | Koslover, D.J, Callaghan, A.J, Marcaida, M.J, Martick, M, Scott, W.G, Luisi, B.F. | | Deposit date: | 2008-01-28 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Crystal Structure of the Escherichia Coli Rnase E Apoprotein and a Mechanism for RNA Degradation.
Structure, 16, 2008
|
|
2VRT
 
 | | Crystal Structure of E. coli RNase E possessing M1 RNA fragments - Catalytic Domain | | Descriptor: | 5'-R(*UP*UP)-3', 5'-R(*UP*UP*GP)-3', RIBONUCLEASE E, ... | | Authors: | Koslover, D.J, Callaghan, A.J, Marcaida, M.J, Garman, E.F, Martick, M, Scott, W.G, Luisi, B.F. | | Deposit date: | 2008-04-14 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Crystal Structure of the Escherichia Coli Rnase E Apoprotein and a Mechanism for RNA Degradation.
Structure, 16, 2008
|
|
285D
 
 | |
2WVH
 
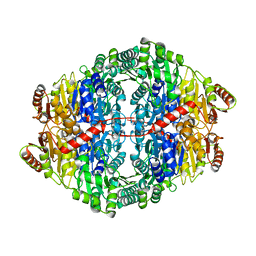 | |
1D98
 
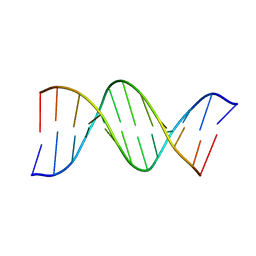 | | THE STRUCTURE OF AN OLIGO(DA).OLIGO(DT) TRACT AND ITS BIOLOGICAL IMPLICATIONS | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*AP*AP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*TP*TP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Nelson, H.C.M, Finch, J.T, Luisi, B.F, Klug, A. | | Deposit date: | 1992-10-17 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of an oligo(dA).oligo(dT) tract and its biological implications.
Nature, 330, 1987
|
|
2CE1
 
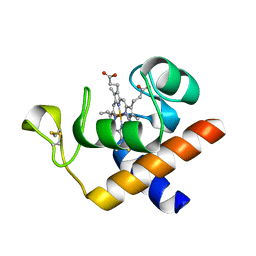 | | Structure of reduced Arabidopsis thaliana cytochrome 6A | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Marcaida, M.J, Schlarb-Ridley, B.G, Worrall, J.A.R, Wastl, J, Evans, T.J, Bendall, D.S, Luisi, B.F, Howe, C.J. | | Deposit date: | 2006-02-02 | | Release date: | 2006-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Cytochrome C(6A), a Novel Dithio-Cytochrome of Arabidopsis Thaliana, and its Reactivity with Plastocyanin: Implications for Function.
J.Mol.Biol., 360, 2006
|
|
2C8M
 
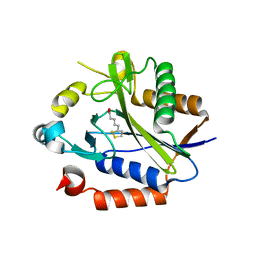 | |
2CE0
 
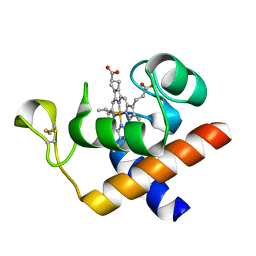 | | Structure of oxidized Arabidopsis thaliana cytochrome 6A | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Marcaida, M.J, Schlarb-Ridley, B.G, Worrall, J.A.R, Wastl, J, Evans, T.J, Bendall, D.S, Luisi, B.F, Howe, C.J. | | Deposit date: | 2006-02-01 | | Release date: | 2006-07-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structure of Cytochrome C(6A), a Novel Dithio-Cytochrome of Arabidopsis Thaliana, and its Reactivity with Plastocyanin: Implications for Function.
J.Mol.Biol., 360, 2006
|
|
2XDB
 
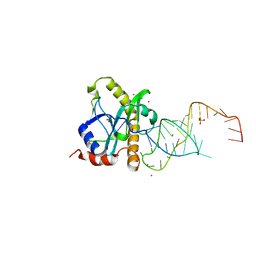 | | A processed non-coding RNA regulates a bacterial antiviral system | | Descriptor: | COBALT (II) ION, SULFATE ION, TOXI, ... | | Authors: | Blower, T.R, Pei, X.Y, Short, F.L, Fineran, P.C, Humphreys, D.P, Luisi, B.F, Salmond, G.P.C. | | Deposit date: | 2010-04-30 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Processed Noncoding RNA Regulates an Altruistic Bacterial Antiviral System.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2AAO
 
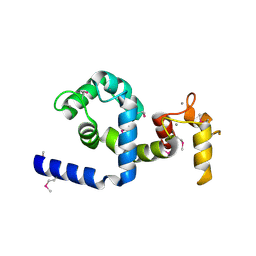 | | Regulatory apparatus of Calcium Dependent protein kinase from Arabidopsis thaliana | | Descriptor: | CALCIUM ION, Calcium-dependent protein kinase, isoform AK1 | | Authors: | Chandran, V, Stollar, E.J, Lindorff-Larsen, K, Harper, J.F, Chazin, W.J, Dobson, C.M, Luisi, B.F, Christodoulou, J. | | Deposit date: | 2005-07-13 | | Release date: | 2005-12-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the regulatory apparatus of a calcium-dependent protein kinase (CDPK): a novel mode of calmodulin-target recognition.
J.Mol.Biol., 357, 2006
|
|
5NB9
 
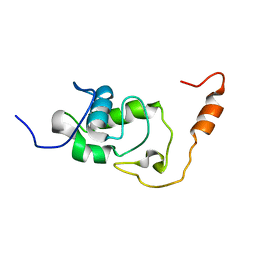 | | Structure of the N-terminal domain of the Escherichia Coli ProQ RNA binding protein | | Descriptor: | RNA chaperone ProQ | | Authors: | Gonzales, G, Hardwick, S, Maslen, S, Skehel, M, Holmqvist, E, Vogel, J, Bateman, A, Luisi, B, Broadhurst, R. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of the Escherichia coli ProQ RNA-binding protein.
RNA, 23, 2017
|
|
5NBB
 
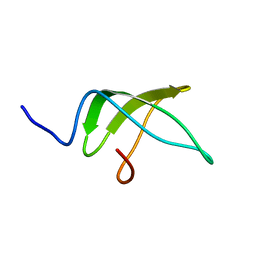 | | Structure of the C-terminal domain of the Escherichia Coli ProQ RNA binding protein | | Descriptor: | RNA chaperone ProQ | | Authors: | Gonzales, G, Hardwick, S, Maslen, S, Skehel, M, Holmqvist, E, Vogel, J, Bateman, A, Luisi, B, Broadhurst, R. | | Deposit date: | 2017-03-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure of the Escherichia coli ProQ RNA-binding protein.
RNA, 23, 2017
|
|
2C7I
 
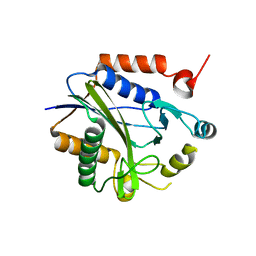 | |
5OHG
 
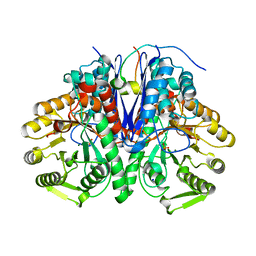 | | enolase in complex with RNase E | | Descriptor: | Enolase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Du, D, Luisi, B.F. | | Deposit date: | 2017-07-16 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Analysis of the natively unstructured RNA/protein-recognition core in the Escherichia coli RNA degradosome and its interactions with regulatory RNA/Hfq complexes.
Nucleic Acids Res., 46, 2018
|
|
5O5S
 
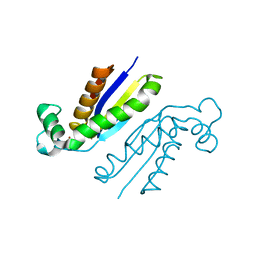 | | X-ray crystal structure of the RapZ C-terminal domain from Escherichia coli | | Descriptor: | MALONATE ION, RNase adapter protein RapZ | | Authors: | Gonzalez, G.M, Durica-Mitic, S, Hardwick, S.W, Moncrieffe, M, Resch, M, Neumann, P, Ficner, R, Gorke, B, Luisi, B.F. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural insights into RapZ-mediated regulation of bacterial amino-sugar metabolism.
Nucleic Acids Res., 45, 2017
|
|
2C0B
 
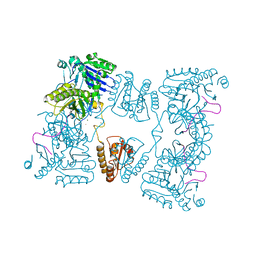 | | Catalytic domain of E. coli RNase E in complex with 13-mer RNA | | Descriptor: | 5'-R(*UP*UP*UP*AP*CP*AP*GP*UP*AP*UP*UP*UP*G)-3', MAGNESIUM ION, RIBONUCLEASE E, ... | | Authors: | Marcaida, M.J, Callaghan, A.J, Scott, W.G, Luisi, B.F. | | Deposit date: | 2005-08-30 | | Release date: | 2005-10-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structure of E. Coli Rnase E Catalytic Domain and Implications for RNA Processing and Turnover
Nature, 437, 2005
|
|
2C4R
 
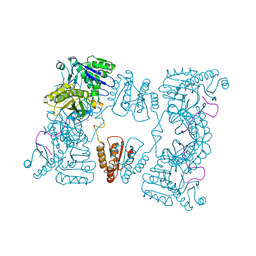 | | Catalytic domain of E. coli RNase E | | Descriptor: | MAGNESIUM ION, RIBONUCLEASE E, SSRNA MOLECULE: 5'-R(*AP*CP*AP*GP*UP*AP*UP*UP*UP*GP)-3', ... | | Authors: | Marcaida, M.J, Callaghan, A.J, Luisi, B.F. | | Deposit date: | 2005-10-21 | | Release date: | 2005-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of E. Coli Rnase E Catalytic Domain and Implications for RNA Processing and Turnover
Nature, 437, 2005
|
|
5O66
 
 | | Asymmetric AcrABZ-TolC | | Descriptor: | Multidrug efflux pump accessory protein AcrZ, Multidrug efflux pump subunit AcrA, Multidrug efflux pump subunit AcrB, ... | | Authors: | Du, D, Luisi, B.F. | | Deposit date: | 2017-06-05 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump.
Elife, 6, 2017
|
|
2BP7
 
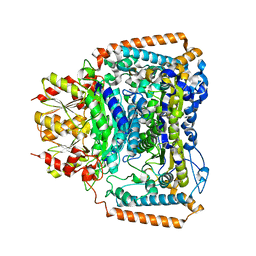 | | New crystal form of the Pseudomonas putida branched-chain dehydrogenase (E1) | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT | | Authors: | Frank, R.A.W, Pratap, J.V, Pei, X.Y, Perham, R.N, Luisi, B.F. | | Deposit date: | 2005-04-18 | | Release date: | 2005-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Molecular Origins of Specificity in the Assembly of a Multienzyme Complex.
Structure, 13, 2005
|
|
