3C0D
 
 | | Crystal structure of the putative nitrite reductase NADPH (small subunit) oxidoreductase protein Q87HB1. Northeast Structural Genomics Consortium target VpR162 | | Descriptor: | Putative nitrite reductase NADPH (Small subunit) oxidoreductase protein | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Vorobiev, S.M, Wang, D, Fang, Y, Owens, L, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-19 | | Release date: | 2008-03-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the putative nitrite reductase NADPH (small subunit) oxidoreductase protein Q87HB1.
To be Published
|
|
3SWV
 
 | | Crystal Structure of a domain of Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (Brefeldin A-inhibited GEP 2) from Homo sapiens (Human), Northeast Structural Genomics Consortium target id HR5562A | | Descriptor: | Brefeldin A-inhibited guanine nucleotide-exchange protein 2 | | Authors: | Seetharaman, J, Su, M, Forouhar, F, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-14 | | Release date: | 2011-08-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a domain of Brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BrefeldinA-inhibited GEP 2) from Homo sapiens (Human), Northeast Structural Genomics Consortium target id HR5562A
To be Published
|
|
5E00
 
 | | Structure of HLA-A2 P130 | | Descriptor: | Beta-2-microglobulin, GLY-VAL-TRP-ILE-ARG-THR-PRO-PRO-ALA, HLA class I histocompatibility antigen, ... | | Authors: | Zhang, Y, Wu, Y, Qi, J, Liu, J, Gao, G.F, Meng, S. | | Deposit date: | 2015-09-26 | | Release date: | 2017-01-18 | | Last modified: | 2019-01-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CD8+T-Cell Response-Associated Evolution of Hepatitis B Virus Core Protein and Disease Progress.
J. Virol., 92, 2018
|
|
4DN5
 
 | | Crystal Structure of NF-kB-inducing Kinase (NIK) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Min, X, Liu, J, Sudom, A, Walker, N.P, Wang, Z. | | Deposit date: | 2012-02-08 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Nuclear Factor Kappa B-inducing kinase domain reveals a constitutively active conformation
J.Biol.Chem., 287, 2012
|
|
2R1L
 
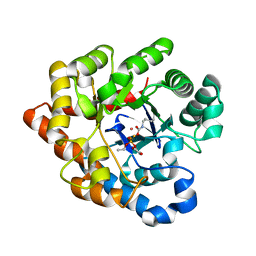 | | OpdA from Agrobacterium radiobacter with bound diethyl thiophosphate from crystal soaking with the compound- 1.95 A | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, FE (II) ION, ... | | Authors: | Ollis, D.L, Jackson, C.J, Foo, J.L, Kim, H.K, Carr, P.D, Liu, J.W, Salem, G. | | Deposit date: | 2007-08-23 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | In crystallo capture of a Michaelis complex and product-binding modes of a bacterial phosphotriesterase
J.Mol.Biol., 375, 2008
|
|
2R1N
 
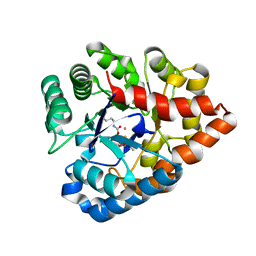 | | OpdA from Agrobacterium radiobacter with bound slow substrate diethyl 4-methoxyphenyl phosphate (20h)- 1.7 A | | Descriptor: | COBALT (II) ION, DIETHYL 4-METHOXYPHENYL PHOSPHATE, FE (II) ION, ... | | Authors: | Ollis, D.L, Jackson, C.J, Foo, J.L, Kim, H.K, Carr, P.D, Liu, J.W, Salem, G. | | Deposit date: | 2007-08-23 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In crystallo capture of a Michaelis complex and product-binding modes of a bacterial phosphotriesterase
J.Mol.Biol., 375, 2008
|
|
2R1P
 
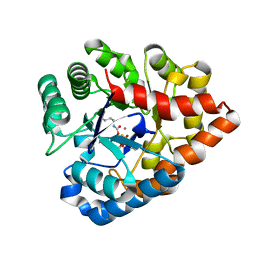 | | OpdA from Agrobacterium radiobacter with bound product diethyl thiophosphate from co-crystallisation with tetraethyl dithiopyrophosphate- 1.8 A | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, FE (II) ION, ... | | Authors: | Ollis, D.L, Jackson, C.J, Foo, J.L, Kim, H.K, Carr, P.D, Liu, J.W, Salem, G. | | Deposit date: | 2007-08-23 | | Release date: | 2008-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In crystallo capture of a Michaelis complex and product-binding modes of a bacterial phosphotriesterase
J.Mol.Biol., 375, 2008
|
|
2QGS
 
 | | Crystal structure of SE1688 protein from Staphylococcus epidermidis. Northeast Structural Genomics Consortium target SeR89 | | Descriptor: | Protein SE1688, ZINC ION | | Authors: | Forouhar, F, Su, M, Benach, J, Seetharaman, J, Chen, C.X, Fang, Y, Cunningham, K, Owen, L, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of SE1688 protein from Staphylococcus epidermidis.
To be Published
|
|
2R1M
 
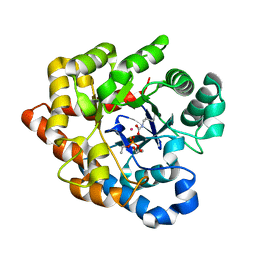 | | OpdA from Agrobacterium radiobacter with bound product diethyl phosphate from crystal soaking with diethyl 4-methoxyphenyl phosphate (450h)- 2.5 A | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, DIETHYL HYDROGEN PHOSPHATE, ... | | Authors: | Ollis, D.L, Jackson, C.J, Foo, J.L, Kim, H.K, Carr, P.D, Liu, J.W, Salem, G. | | Deposit date: | 2007-08-23 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | In crystallo capture of a Michaelis complex and product-binding modes of a bacterial phosphotriesterase
J.Mol.Biol., 375, 2008
|
|
2QGN
 
 | | Crystal structure of tRNA isopentenylpyrophosphate transferase (BH2366) from Bacillus halodurans, Northeast Structural Genomics Consortium target BhR41. | | Descriptor: | SULFATE ION, tRNA delta(2)-isopentenylpyrophosphate transferase | | Authors: | Forouhar, F, Neely, H, Abashidze, M, Seetharaman, J, Shastry, R, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of tRNA isopentenylpyrophosphate transferase (BH2366) from Bacillus halodurans.
To be Published
|
|
5WE0
 
 | | Structural Basis for Shelterin Bridge Assembly | | Descriptor: | DNA-binding protein rap1, Protection of telomeres protein poz1, Protection of telomeres protein tpz1, ... | | Authors: | Kim, J.-K, Liu, J, Hu, X, Yu, C, Roskamp, K, Sankaran, B, Huang, L, Komives, E.-A, Qiao, F. | | Deposit date: | 2017-07-06 | | Release date: | 2017-12-20 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Shelterin Bridge Assembly.
Mol. Cell, 68, 2017
|
|
2QGZ
 
 | | Crystal structure of a putative primosome component from Streptococcus pyogenes serotype M3. Northeast Structural Genomics target DR58 | | Descriptor: | PHOSPHATE ION, Putative primosome component | | Authors: | Seetharaman, J, Chen, Y, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xia, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of a putative primosome component from Streptococcus pyogenes serotype M3.
To be Published
|
|
5WE1
 
 | | Structural Basis for Shelterin Bridge Assembly | | Descriptor: | Protection of telomeres protein poz1,Protection of telomeres protein poz1, Protection of telomeres protein tpz1, ZINC ION | | Authors: | Kim, J.-K, Liu, J, Hu, X, Yu, C, Roskamp, K, Sankaran, B, Huang, L, Komives, E.-A, Qiao, F. | | Deposit date: | 2017-07-06 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural Basis for Shelterin Bridge Assembly.
Mol. Cell, 68, 2017
|
|
6U7V
 
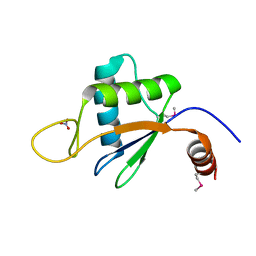 | | xRRM structure of spPof8 | | Descriptor: | NITRATE ION, Protein pof8 | | Authors: | Kim, J.-K, Hu, X, Yu, C, Jun, H.-I, Liu, J, Sankaran, B, Huang, L, Qiao, F. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Quality-Control Mechanism for Telomerase RNA Folding in the Cell.
Cell Rep, 33, 2020
|
|
2QGU
 
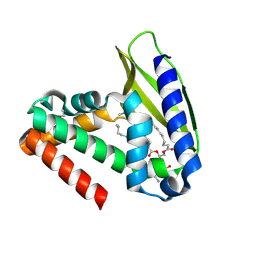 | | Three-dimensional structure of the phospholipid-binding protein from Ralstonia solanacearum Q8XV73_RALSQ in complex with a phospholipid at the resolution 1.53 A. Northeast Structural Genomics Consortium target RsR89 | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Probable signal peptide protein | | Authors: | Kuzin, A.P, Chen, Y, Jayaraman, S, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-dimensional structure of the phospholipid-binding protein from Ralstonia solanacearum Q8XV73_RALSQ in complex with a phospholipid at the resolution 1.53 A.
To be Published
|
|
5WZT
 
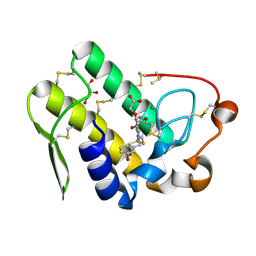 | | Crystal structure of human secreted phospholipase A2 group IIE with Compound 14 | | Descriptor: | 2-[1-[(3-bromophenyl)methyl]-2-methyl-3-oxamoyl-indol-4-yl]oxyethanoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Hou, S, Xu, J, Xu, T, Liu, J. | | Deposit date: | 2017-01-18 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for functional selectivity and ligand recognition revealed by crystal structures of human secreted phospholipase A2 group IIE
Sci Rep, 7, 2017
|
|
2QGM
 
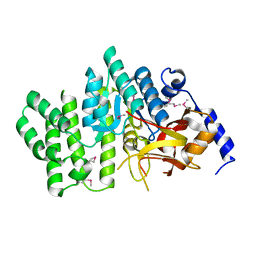 | | Crystal structure of succinoglycan biosynthesis protein at the resolution 1.7 A. Northeast Structural Genomics Consortium target BcR136. | | Descriptor: | Succinoglycan biosynthesis protein | | Authors: | Kuzin, A.P, Abashidze, M, Jayaraman, S, Wang, H, Fang, Y, Maglaqui, M, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-24 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of succinoglycan biosynthesis protein at the resolution 1.7 A. Northeast Structural Genomics Consortium target BcR136.
To be Published
|
|
6DRZ
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8alpha)-N-[(2S)-1-hydroxybutan-2-yl]-1,6-dimethyl-9,10-didehydroergoline-8-carboxamide, 5HT2B receptor, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DRY
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (8beta)-N-[(2S)-1-hydroxybutan-2-yl]-6-methyl-9,10-didehydroergoline-8-carboxamide, 5HT2B receptor, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.918 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DRX
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5HT2B receptor, BRIL chimera, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DS0
 
 | | Structural Determinants of Activation and Biased Agonism at the 5-HT2B Receptor | | Descriptor: | (1S)-1-[(2-chloro-3,4-dimethoxyphenyl)methyl]-6-methyl-2,3,4,9-tetrahydro-1H-beta-carboline, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5HT2B receptor, ... | | Authors: | McCorvy, J.D, Wacker, D, Wang, S, Agegnehu, B, Liu, J, Lansu, K, Tribo, A.R, Olsen, R.H.J, Che, T, Jin, J, Roth, B.L. | | Deposit date: | 2018-06-13 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.188 Å) | | Cite: | Structural determinants of 5-HT2Breceptor activation and biased agonism.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4USG
 
 | | Crystal structure of PC4 W89Y mutant complex with DNA | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP *TP*TP*TP*TP*TP*G)-3', ACTIVATED RNA POLYMERASE II TRANSCRIPTIONAL COACTIVATOR P15 | | Authors: | Zhao, Y, Liu, J. | | Deposit date: | 2014-07-08 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.973 Å) | | Cite: | Substitution of Tryptophan 89 with Tyrosine Switches the DNA Binding Mode of Pc4.
Sci.Rep., 5, 2015
|
|
7QUR
 
 | | SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C3 symmetry | | Descriptor: | 2,3,5-tris(iodanyl)benzamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-01 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
7QUS
 
 | | SARS-CoV-2 Spike, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-08 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
5EP6
 
 | | The crystal structure of NAP1 in complex with TBK1 | | Descriptor: | 5-azacytidine-induced protein 2, GLYCEROL, Serine/threonine-protein kinase TBK1 | | Authors: | Li, F, Xie, X, Liu, J, Pan, L. | | Deposit date: | 2015-11-11 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins.
Nat Commun, 7, 2016
|
|
