2DCI
 
 | |
5JHD
 
 | | Crystal structure of LS10-TCR/M1-HLA-A*02 complex | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Stern, L.J, Selin, L.K, Song, I. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Broad TCR repertoire and diverse structural solutions for recognition of an immunodominant CD8(+) T cell epitope.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5ISZ
 
 | | Crystal structure of LS01-TCR/M1-HLA-A*02 complex | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Stern, L.J, Selin, L.K, Song, I. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Broad TCR repertoire and diverse structural solutions for recognition of an immunodominant CD8(+) T cell epitope.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5I5I
 
 | |
2BLG
 
 | | STRUCTURAL BASIS OF THE TANFORD TRANSITION OF BOVINE BETA-LACTOGLOBULIN FROM CRYSTAL STRUCTURES AT THREE PH VALUES; PH 8.2 | | Descriptor: | BETA-LACTOGLOBULIN | | Authors: | Qin, B.Y, Bewley, M.C, Creamer, L.K, Baker, H.M, Baker, E.N, Jameson, G.B. | | Deposit date: | 1998-08-29 | | Release date: | 1999-01-27 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis of the Tanford transition of bovine beta-lactoglobulin.
Biochemistry, 37, 1998
|
|
5I5M
 
 | | Shewanella denitrificans nitrous oxide reductase, Ca2+-reconstituted form | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CALCIUM ION, Nitrous-oxide reductase, ... | | Authors: | Schneider, L.K, Einsle, O. | | Deposit date: | 2016-02-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Role of Calcium in Secondary Structure Stabilization during Maturation of Nitrous Oxide Reductase.
Biochemistry, 55, 2016
|
|
5LAR
 
 | | Crystal structure of p38 alpha MAPK14 in complex with VPC00628 | | Descriptor: | 5-azanyl-~{N}-[[4-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, Mitogen-activated protein kinase 14 | | Authors: | Chaikuad, A, Petersen, L.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-14 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel p38alpha MAP kinase inhibitors identified from yoctoReactor DNA-encoded small molecule library
Medchemcomm, 7, 2016
|
|
2FAO
 
 | | Crystal Structure of Pseudomonas aeruginosa LigD polymerase domain | | Descriptor: | SULFATE ION, probable ATP-dependent DNA ligase | | Authors: | Zhu, H, Nandakumar, J, Aniukwu, J, Wang, L.K, Glickman, M.S, Lima, C.D, Shuman, S. | | Deposit date: | 2005-12-07 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Atomic structure and nonhomologous end-joining function of the polymerase component of bacterial DNA ligase D
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2FAQ
 
 | | Crystal Structure of Pseudomonas aeruginosa LigD polymerase domain with ATP and Manganese | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Zhu, H, Nandakumar, J, Aniukwu, J, Wang, L.K, Glickman, M.S, Lima, C.D, Shuman, S. | | Deposit date: | 2005-12-07 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Atomic structure and nonhomologous end-joining function of the polymerase component of bacterial DNA ligase D
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2FAR
 
 | | Crystal Structure of Pseudomonas aeruginosa LigD polymerase domain with dATP and Manganese | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Zhu, H, Nandakumar, J, Aniukwu, J, Wang, L.K, Glickman, M.S, Lima, C.D, Shuman, S. | | Deposit date: | 2005-12-07 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Atomic structure and nonhomologous end-joining function of the polymerase component of bacterial DNA ligase D
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2GDV
 
 | |
2Z31
 
 | | Crystal structure of immune receptor complex | | Descriptor: | H-2 class II histocompatibility antigen, A-U alpha chain, A-U beta chain precursor, ... | | Authors: | Feng, D, Bond, C.J, Ely, L.K, Garcia, K.C. | | Deposit date: | 2007-05-30 | | Release date: | 2007-10-09 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction 'codon'.
Nat.Immunol., 8, 2007
|
|
4CPH
 
 | | trans-divalent streptavidin with love-hate ligand 4 | | Descriptor: | 5-[(3aS,4S,6aR)-2-oxo-hexahydro-1H-thieno[3,4- d]imidazolidin-4-yl]-N'-{2,6-bis[4-(morpholine-4- sulfonyl)phenyl]phenyl}pentanehydrazide, STREPTAVIDIN | | Authors: | Fairhead, M, Shen, D, Chan, L.K.M, Lowe, E.D, Donohoe, T.J, Howarth, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Love-Hate Ligands for High Resolution Analysis of Strain in Ultra-Stable Protein/Small Molecule Interaction.
Bioorg.Med.Chem., 22, 2014
|
|
4CPI
 
 | | streptavidin A86D mutant with love-hate ligand 4 | | Descriptor: | 5-[(3aS,4S,6aR)-2-oxo-hexahydro-1H-thieno[3,4- d]imidazolidin-4-yl]-N'-{2,6-bis[4-(morpholine-4- sulfonyl)phenyl]phenyl}pentanehydrazide, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fairhead, M, Shen, D, Chan, L.K.M, Lowe, E.D, Donohoe, T.J, Howarth, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Love-Hate Ligands for High Resolution Analysis of Strain in Ultra-Stable Protein/Small Molecule Interaction.
Bioorg.Med.Chem., 22, 2014
|
|
4CPF
 
 | | Wild-type streptavidin in complex with love-hate ligand 3 (LH3) | | Descriptor: | STREPTAVIDIN, methyl 4-(2-{5-[(3aS,4S,6aR)-2-oxo-hexahydro-1H- thieno[3,4-d]imidazolidin-4-yl]pentanehydrazido}-3- [4-(methoxycarbonyl)phenyl]phenyl)benzoate | | Authors: | Fairhead, M, Shen, D, Chan, L.K.M, Lowe, E.D, Donohoe, T.J, Howarth, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Love-Hate Ligands for High Resolution Analysis of Strain in Ultra-Stable Protein/Small Molecule Interaction.
Bioorg.Med.Chem., 22, 2014
|
|
4CPE
 
 | | Wild-type streptavidin in complex with love-hate ligand 1 (LH1) | | Descriptor: | (3aS,4S,6aR)-2-oxo-hexahydro-1H-thieno[3,4- d]imidazolidin-4-yl]-N-{2-[(2,6- diphenylphenyl)formamido]ethyl}pentanamide, STREPTAVIDIN | | Authors: | Fairhead, M, Shen, D, Chan, L.K.M, Lowe, E.D, Donohoe, T.J, Howarth, M. | | Deposit date: | 2014-02-06 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Love-Hate Ligands for High Resolution Analysis of Strain in Ultra-Stable Protein/Small Molecule Interaction.
Bioorg.Med.Chem., 22, 2014
|
|
8Q3E
 
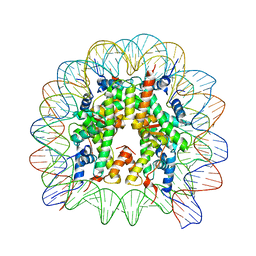 | | High Resolution Structure of Nucleosome Core with Bound Foamy Virus GAG Peptide | | Descriptor: | DNA (145-MER), GLY-GLY-TYR-ASN-LEU-ARG-PRO-ARG-THR-TYR-GLN-PRO-GLN-ARG-TYR-GLY-GLY-GLY, Histone H2A type 1-B/E, ... | | Authors: | De Falco, L, Batchelor, L.K, Dyson, P.J, Davey, C.A. | | Deposit date: | 2023-08-04 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Viral peptide conjugates for metal-warhead delivery to chromatin.
Rsc Adv, 14, 2024
|
|
8Q3M
 
 | | Structure of Nucleosome Core with a Bound Kaposi Sarcoma Associated Herpesvirus LANA Peptide Having a Methionine to Ornithine Substitution | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | De Falco, L, Batchelor, L.K, Dyson, P.J, Davey, C.A. | | Deposit date: | 2023-08-04 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Viral peptide conjugates for metal-warhead delivery to chromatin.
Rsc Adv, 14, 2024
|
|
8Q36
 
 | | Structure of Nucleosome Core with a Bound Metallopeptide Conjugate (Foamy Virus GAG Peptide-Au[I] Compound) | | Descriptor: | DNA (145-MER), GAG structural protein, Histone H2A type 1-B/E, ... | | Authors: | De Falco, L, Batchelor, L.K, Dyson, P.J, Davey, C.A. | | Deposit date: | 2023-08-03 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Viral peptide conjugates for metal-warhead delivery to chromatin.
Rsc Adv, 14, 2024
|
|
8Q3X
 
 | | Structure of Nucleosome Core with a Bound Metallopeptide Conjugate (Kaposi Sarcoma Associated Herpesvirus LANA Peptide-Au[I] Compound) | | Descriptor: | 4-diphenylphosphanylbenzoic acid, DNA (145-MER), GOLD ION, ... | | Authors: | De Falco, L, Batchelor, L.K, Dyson, P.J, Davey, C.A. | | Deposit date: | 2023-08-04 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Viral peptide conjugates for metal-warhead delivery to chromatin.
Rsc Adv, 14, 2024
|
|
3BZF
 
 | | The human non-classical major histocompatibility complex molecule HLA-E | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, alpha chain E, ... | | Authors: | Hoare, H.L, Sullivan, L.C, Ely, L.K, Beddoe, T, Henderson, K.N, Lin, J, Clements, C.S, Reid, H.H, Brooks, A.G, Rossjohn, J. | | Deposit date: | 2008-01-17 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Subtle changes in peptide conformation profoundly affect recognition of the non-classical MHC class I molecule HLA-E by the CD94-NKG2 natural killer cell receptors
J.Mol.Biol., 377, 2008
|
|
4CZS
 
 | | Discovery of Glycomimetic Ligands via Genetically-encoded Library of Phage displaying Mannose-peptides | | Descriptor: | 2-hydroxyethyl alpha-D-mannopyranoside, CALCIUM ION, Concanavalin V, ... | | Authors: | Ng, S, Lin, E, Tjhung, K.F, Gerlits, O, Sood, A, Kasper, B, Deng, L, Kitov, P.I, Matochko, W.L, Paschal, B.M, Noren, C.J, Klassen, J, Mahal, L.K, Coates, L, Woods, R.J, Derda, R. | | Deposit date: | 2014-04-22 | | Release date: | 2015-04-22 | | Last modified: | 2022-12-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Genetically-Encoded Fragment-Based Discovery of Glycopeptide Ligands for Carbohydrate-Binding Proteins.
J.Am.Chem.Soc., 137, 2015
|
|
3BXR
 
 | | Crystal Structures Of Highly Constrained Substrate And Hydrolysis Products Bound To HIV-1 Protease. Implications For Catalytic Mechanism | | Descriptor: | (9S,12S)-9-(1-methylethyl)-N-[(8S,11S)-8-[(1S)-1-methylpropyl]-7,10-dioxo-2-oxa-6,9-diazabicyclo[11.2.2]heptadeca-1(15),13,16-trien-11-yl]-7,10-dioxo-2-oxa-8,11-diazabicyclo[12.2.2]octadeca-1(16),14,17-triene-12-carboxamide, Protease, SULFATE ION | | Authors: | Tyndall, J.D, Pattenden, L.K, Reid, R.C, Hu, S.H, Alewood, D, Alewood, P.F, Walsh, T, Fairlie, D.P, Martin, J.L. | | Deposit date: | 2008-01-14 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Highly Constrained Substrate and Hydrolysis Products Bound to HIV-1 Protease. Implications for the Catalytic Mechanism
Biochemistry, 47, 2008
|
|
8QA4
 
 | | MTHFR + SAH symmetric dis-inhibited state | | Descriptor: | Methylenetetrahydrofolate reductase (NADPH), S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Blomgren, L.K.M, Yue, W.W, Froese, D.S, McCorvie, T.J. | | Deposit date: | 2023-08-22 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Dynamic inter-domain transformations mediate the allosteric regulation of human 5, 10-methylenetetrahydrofolate reductase.
Nat Commun, 15, 2024
|
|
8QA6
 
 | | MTHFR + SAM inhibited state | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methylenetetrahydrofolate reductase (NADPH), S-ADENOSYLMETHIONINE | | Authors: | Blomgren, L.K.M, Yue, W.W, Froese, D.S, McCorvie, T.J. | | Deposit date: | 2023-08-22 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Dynamic inter-domain transformations mediate the allosteric regulation of human 5, 10-methylenetetrahydrofolate reductase.
Nat Commun, 15, 2024
|
|
