7JFQ
 
 | | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, FORMIC ACID | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-17 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of 3CL MainPro of SARS-CoV-2 with de-oxidized C145
To Be Published
|
|
1UW3
 
 | | The crystal structure of the globular domain of sheep prion protein | | Descriptor: | GLUTATHIONE, PHOSPHATE ION, PRION PROTEIN | | Authors: | Haire, L.F, Whyte, S.M, Vasisht, N, Gill, A.C, Verma, C, Dodson, E.J, Dodson, G.G, Bayley, P.M. | | Deposit date: | 2004-01-29 | | Release date: | 2004-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Crystal Structure of the Globular Domain of Sheep Prion Protein
J.Mol.Biol., 336, 2004
|
|
1UN8
 
 | | Crystal structure of the dihydroxyacetone kinase of C. freundii (native form) | | Descriptor: | (2R)-3-(PHOSPHONOOXY)-2-(TETRADECANOYLOXY)PROPYL PALMITATE, DIHYDROXYACETONE KINASE | | Authors: | Siebold, C, Arnold, I, Garcia-Alles, L.F, Baumann, U, Erni, B. | | Deposit date: | 2003-09-08 | | Release date: | 2003-10-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Citrobacter Freundii Dihydroxyacetone Kinase Reveals an Eight-Stranded Alpha-Helical Barrel ATP-Binding Domain
J.Biol.Chem., 278, 2003
|
|
1UOE
 
 | | Crystal structure of the dihydroxyacetone kinase from E. coli in complex with glyceraldehyde | | Descriptor: | DIHYDROXYACETONE KINASE, GLYCEROL, SULFATE ION | | Authors: | Siebold, C, Garcia-Alles, L.F, Luthi-Nyffeler, T, Flukiger-Bruhwiler, K, Burgi, H.-B, Baumann, U, Erni, B. | | Deposit date: | 2003-09-16 | | Release date: | 2004-09-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphoenolpyruvate- and ATP-Dependent Dihydroxyacetone Kinases: Covalent Substrate-Binding and Kinetic Mechanism
Biochemistry, 43, 2004
|
|
5ABM
 
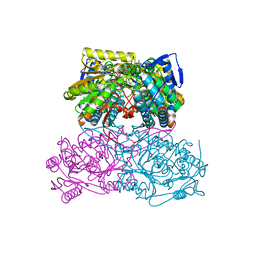 | | Sheep aldehyde dehydrogenase 1A1 | | Descriptor: | MAGNESIUM ION, RETINAL DEHYDROGENASE 1, [[(2R,3S,4R,5R)-5-[(3R)-3-aminocarbonyl-3,4-dihydro-2H-pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanidyl-ph osphoryl] [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl phosphate | | Authors: | Koch, M.F, Harteis, S, Blank, I.D, Pestel, G, Tietze, L.F, Ochsenfeld, C, Schneider, S, Sieber, S.A. | | Deposit date: | 2015-08-07 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, Biochemical, and Computational Studies Reveal the Mechanism of Selective Aldehyde Dehydrogenase 1A1 Inhibition by Cytotoxic Duocarmycin Analogues.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7JU7
 
 | | The crystal structure of SARS-CoV-2 Main Protease in complex with masitinib | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-19 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2.
Science, 373, 2021
|
|
7KGJ
 
 | | Crystal structure of synthetic nanobody (Sb45) complexes with SARS-CoV-2 receptor binding domain | | Descriptor: | Sb45, Sybody-45, Synthetic Nanobody, ... | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2020-10-16 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
7KLW
 
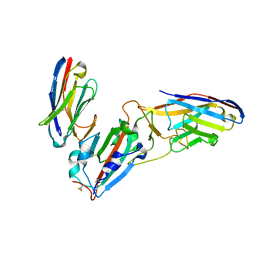 | | Crystal structure of synthetic nanobody (Sb45+Sb68) complexes with SARS-CoV-2 receptor binding domain | | Descriptor: | SB45, Synthetic Nanobody, SB68, ... | | Authors: | Jiang, J, Ahmad, J, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2020-11-01 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
5HLJ
 
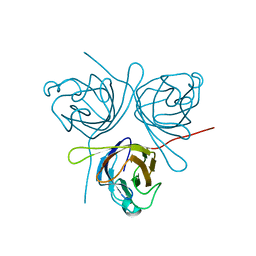 | | Crystal Structure of Major Envelope Protein VP24 from White Spot Syndrome Virus | | Descriptor: | VP24 | | Authors: | Sun, L.F, Su, Y.T, Zhao, Y.H, Fu, Z.Q, Wu, Y.K. | | Deposit date: | 2016-01-15 | | Release date: | 2016-09-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Crystal Structure of Major Envelope Protein VP24 from White Spot Syndrome Virus
Sci Rep, 6, 2016
|
|
5AC1
 
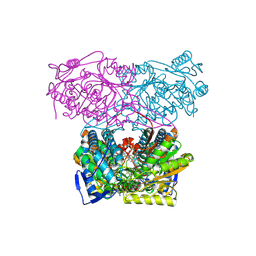 | | Sheep aldehyde dehydrogenase 1A1 with duocarmycin analog inhibitor | | Descriptor: | 1-[(1S)-1-methyl-5-oxidanyl-1,2-dihydrobenzo[e]indol-3-yl]hexan-1-one, MAGNESIUM ION, RETINAL DEHYDROGENASE 1, ... | | Authors: | Koch, M.F, Harteis, S, Blank, I.D, Pestel, G, Tietze, L.F, Ochsenfeld, C, Schneider, S, Sieber, S.A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural, Biochemical, and Computational Studies Reveal the Mechanism of Selective Aldehyde Dehydrogenase 1A1 Inhibition by Cytotoxic Duocarmycin Analogues.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5AC0
 
 | | ovis aries Aldehyde Dehydrogenase 1A1 in complex with a duocarmycin analog | | Descriptor: | 1-[(1S)-1-methyl-5-oxidanyl-1,2-dihydrobenzo[e]indol-3-yl]hexan-1-one, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Koch, M.F, Harteis, S, Blank, I.D, Pestel, G, Tietze, L.F, Ochsenfeld, C, Schneider, S, Sieber, S.A. | | Deposit date: | 2015-08-10 | | Release date: | 2015-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural, Biochemical, and Computational Studies Reveal the Mechanism of Selective Aldehyde Dehydrogenase 1A1 Inhibition by Cytotoxic Duocarmycin Analogues.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7KI3
 
 | |
1UN9
 
 | | Crystal structure of the dihydroxyacetone kinase from C. freundii in complex with AMP-PNP and Mg2+ | | Descriptor: | DIHYDROXYACETONE, DIHYDROXYACETONE KINASE, MAGNESIUM ION, ... | | Authors: | Siebold, C, Arnold, I, Garcia-Alles, L.F, Baumann, U, Erni, B. | | Deposit date: | 2003-09-08 | | Release date: | 2003-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of the Citrobacter Freundii Dihydroxyacetone Kinase Reveals an Eight-Stranded Alpha-Helical Barrel ATP-Binding Domain
J.Biol.Chem., 278, 2003
|
|
3EXR
 
 | | Crystal structure of KGPDC from Streptococcus mutans | | Descriptor: | RmpD (Hexulose-6-phosphate synthase) | | Authors: | Li, G.L, Liu, X, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-16 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
6LHT
 
 | | Localized reconstruction of coxsackievirus A16 mature virion in complex with Fab 18A7 | | Descriptor: | SPHINGOSINE, VP1 protein, heavy chain variable region of Fab 18A7, ... | | Authors: | He, M.Z, Xu, L.F, Zheng, Q.B, Zhu, R, Yin, Z.C, Cheng, T, Li, S.W. | | Deposit date: | 2019-12-10 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Identification of Antibodies with Non-overlapping Neutralization Sites that Target Coxsackievirus A16.
Cell Host Microbe, 27, 2020
|
|
6L72
 
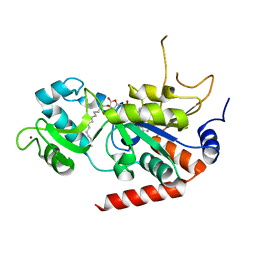 | | Sirtuin 2 demyristoylation native final product | | Descriptor: | NAD-dependent protein deacetylase sirtuin-2, ZINC ION, [(2S,3R,4R,5R)-5-[[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-2,4-bis(oxidanyl)oxolan-3-yl] tetradecanoate | | Authors: | Chen, L.F. | | Deposit date: | 2019-10-30 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Sirtuin 2 protein with H3K18 myristoylated peptide
To Be Published
|
|
6L71
 
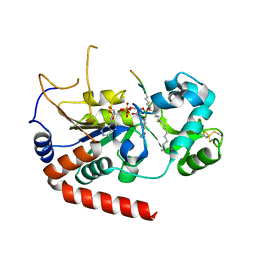 | |
3O5S
 
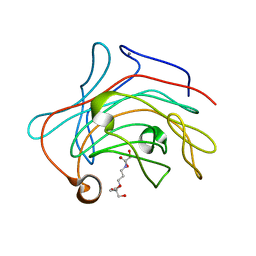 | | Crystal Structure of the endo-beta-1,3-1,4 glucanase from Bacillus subtilis (strain 168) | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase, CALCIUM ION | | Authors: | Santos, C.R, Tonoli, C.C.C, Souza, A.R, Furtado, G.P, Ribeiro, L.F, Ward, R.J, Murakami, M.T. | | Deposit date: | 2010-07-28 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and structural characterization of a Beta-1,3 1,4-glucanase from Bacillus subtilis 168
PROCESS BIOCHEM, 46, 2011
|
|
7PTV
 
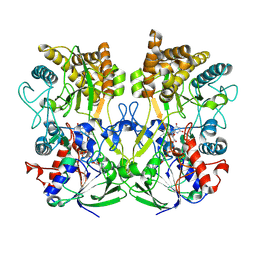 | | Structure of the Mimivirus genomic fibre asymmetric unit | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
6LR3
 
 | | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum | | Descriptor: | Macrophage migration inhibitory factor, SULFATE ION | | Authors: | Su, Z.M, Tian, X.Y, Li, H.J, Wei, Z.M, Chen, L.F, Ren, H.X, Peng, W.F, Tang, C.T. | | Deposit date: | 2020-01-15 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum.
Biochem.J., 477, 2020
|
|
1M8Q
 
 | | Molecular Models of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle | | Descriptor: | Skeletal muscle Actin, Skeletal muscle Myosin II, Skeletal muscle Myosin II Essential Light Chain, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-07-25 | | Release date: | 2002-09-10 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular modeling of averaged rigor crossbridges from tomograms of insect flight muscle.
J.Struct.Biol., 138, 2002
|
|
7T7W
 
 | |
3EXS
 
 | | Crystal structure of KGPDC from Streptococcus mutans in complex with D-R5P | | Descriptor: | RIBULOSE-5-PHOSPHATE, RmpD (Hexulose-6-phosphate synthase) | | Authors: | Li, G.L, Liu, X, Wang, K.T, Li, L.F, Su, X.D. | | Deposit date: | 2008-10-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Open-closed conformational change revealed by the crystal structures of 3-keto-L-gulonate 6-phosphate decarboxylase from Streptococcus mutans
Biochem.Biophys.Res.Commun., 381, 2009
|
|
3H6X
 
 | |
6LKW
 
 | | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum | | Descriptor: | CHLORIDE ION, Macrophage migration inhibitory factor | | Authors: | Su, Z.M, Tian, X.Y, Li, H.J, Wei, Z.M, Chen, L.F, Ren, H.X, Peng, W.F, Tang, C.T. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum.
Biochem.J., 477, 2020
|
|
